Abstract
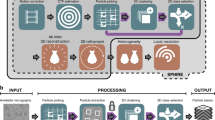
The dramatic growth in the use of cryo-electron microscopy (cryo-EM) to generate high-resolution structures of macromolecular complexes has changed the landscape of structural biology. The majority of structures deposited in the Electron Microscopy Data Bank (EMDB) at higher than 4-Å resolution were collected on Titan Krios microscopes. Although the pipeline for single-particle data collection is becoming routine, there is much variation in how sessions are set up. Furthermore, when collection is under way, there are a range of approaches for efficiently moving and pre-processing these data. Here, we present a standard operating procedure for single-particle data collection with Thermo Fisher Scientific EPU software, using the two most common direct electron detectors (the Thermo Fisher Scientific Falcon 3 (F3EC) and the Gatan K2), as well as a strategy for structuring these data to enable efficient pre-processing and on-the-fly monitoring of data collection. This protocol takes 3–6 h to set up a typical automated data collection session.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cheng, Y., Grigorieff, N., Penczek, P. A. & Walz, T. A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015).
Renaud, J.-P. et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 (2018).
Thompson, R. F., Walker, M., Siebert, C. A., Muench, S. P. & Ranson, N. A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods 100, 3–15 (2016).
Kuehlbrandt, W. The resolution revolution. Science 343, 1443–1444 (2014).
Ruskin, R. S., Yu, Z. & Grigorieff, N. Quantitative characterization of electron detectors for transmission electron microscopy. J. Struct. Biol. 184, 385–393 (2013).
Kuijper, M. et al. FEI’s direct electron detector developments: embarking on a revolution in cryo-TEM. J. Struct. Biol. 192, 179–187 (2015).
McMullan, G., Faruqi, A. R., Clare, D. & Henderson, R. Comparison of optimal performance at 300 keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy 147, 156–163 (2014).
Patwardhan, A. Trends in the Electron Microscopy Data Bank (EMDB). Acta Crystallogr. D Struct. Biol. 73, 503–508 (2017).
Noble, A. J. et al. Routine single particle CryoEM sample and grid characterization by tomography. Elife 7, 32 (2018).
Drulyte, I. et al. Approaches to altering particle distributions in cryo-electron microscopy sample preparation. Acta Crystallogr. D Struct. Biol. 74, 560–571 (2018).
Amporndanai, K. et al. X-ray and cryo-EM structures of inhibitor-bound cytochrome bc1 complexes for structure-based drug discovery. IUCrJ. 5, 200–210 (2018).
Meshcheriakova, Y., Durrant, A., Hesketh, E. L., Ranson, N. A. & Lomonossoff, G. P. Combining high-resolution cryo-electron microscopy and mutagenesis to develop cowpea mosaic virus for bionanotechnology. Biochem. Soc. Trans. 45, 1263–1269 (2017).
Rawson, S. et al. Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM. Proc. Natl. Acad. Sci.USA 115, 1795–1800 (2018).
Baggen, J. et al. Role of enhanced receptor engagement in the evolution of a pandemic acute hemorrhagic conjunctivitis virus. Proc. Natl. Acad. Sci.USA 115, 397–402 (2018).
Scheres, S. H. W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
la Rosa-Trevin de, J. M. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016).
Biyani, N. et al. Focus: the interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife 4, e06980 (2015).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Russo, C. J. & Passmore, L. A. Electron microscopy. Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014).
Hesketh, E. L. et al. The 3.3 Å structure of a plant geminivirus using cryo-EM. Nat. Commun. 9, 2369 (2018).
Iadanza, M. G. et al. The structure of a β2-microglobulin fibril suggests a molecular basis for its amyloid polymorphism. Nat. Commun. 9, 4517 (2018).
Hurdiss, D. L., Frank, M., Snowden, J. S., Macdonald, A. & Ranson, N. A. The structure of an infectious human polyomavirus and its interactions with cellular receptors. Structure 26, 839–847.e3 (2018).
Plaschka, C., Lin, P.-C., Charenton, C. & Nagai, K. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 559, 419–422 (2018).
Conley, M. J. et al. Calicivirus VP2 forms a portal to mediate endosome escape. Preprint at https://www.biorxiv.org/content/early/2018/08/23/397901 (2018).
Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. & Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. 111, 15635–15640 (2014).
Acknowledgements
The Titan Krios microscopes were funded by the University of Leeds (UoL ABSL award) and the Wellcome Trust (108466/Z/15/Z). We are grateful to the EM community at Leeds and our external users for their feedback on our procedures and for the example data collection parameters shown in Table 1. We thank the Faculty of Biological sciences IT team at UoL, in particular P. Pelliccia, A. Richmond and M. Beck, for help with setting up and maintaining the data processing and storage servers and data transfer scripts. E.L.H. is partially funded by BBSRC (BB/L021250/1). M.G.I. received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) ERC grant agreement no. 322408 and the MRC (MR/P018491/1). The EPA_CC_threshold.py script is a modified version of a script kindly provided by R. Danev, who we also thank for helpful discussions about optimal use of the phase plate.
Author information
Authors and Affiliations
Contributions
R.F.T. and E.L.H. wrote the EPU setup protocol. M.G.I. and S.R. wrote the scripts. R.F.T., E.L.H., M.G.I., S.R. and N.A.R. contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Hesketh, E. L. et al. Nat. Commun. 9, 2369 (2018): https://www.nature.com/articles/s41467-018-04793-6
Baggen, J. et al. Proc. Natl. Acad. Sci. USA 115, 397–402 (2018): http://www.pnas.org/content/115/2/397
Agip, A.-N. A. et al. Nat. Struct. Mol. Biol. 25, 548–556 (2018): https://www.nature.com/articles/s41594-018-0073-1
Integrated supplementary information
Supplementary Figure 1
Micrograph analysis output associated with example data (EMPAIR-10205).
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Methods 1–3, Supplementary Notes 1 and 2, and Supplementary Tables 1–4
Supplementary Video 1
Steps 1–11 of the protocol, clipping grids for loading into a Thermo Fisher Scientific autoloader microscope
Rights and permissions
About this article
Cite this article
Thompson, R.F., Iadanza, M.G., Hesketh, E.L. et al. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat Protoc 14, 100–118 (2019). https://doi.org/10.1038/s41596-018-0084-8
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0084-8
This article is cited by
-
Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain
Nature Communications (2024)
-
Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis
Nature Microbiology (2024)
-
Anaerobic cryoEM protocols for air-sensitive nitrogenase proteins
Nature Protocols (2024)
-
Architecture of symbiotic dinoflagellate photosystem I–light-harvesting supercomplex in Symbiodinium
Nature Communications (2024)
-
Insights into the modulation of bacterial NADase activity by phage proteins
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.