Abstract
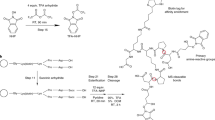
Cross-linking mass spectrometry (XL-MS) has received considerable interest, owing to its potential to investigate protein–protein interactions (PPIs) in an unbiased fashion in complex protein mixtures. Recent developments have enabled the detection of thousands of PPIs from a single experiment. A unique strength of XL-MS, in comparison with other methods for determining PPIs, is that it provides direct spatial information for the detected interactions. This is accomplished by the use of bifunctional cross-linking molecules that link two amino acids in close proximity with a covalent bond. Upon proteolytic digestion, this results in two newly linked peptides, which are identifiable by MS. XL-MS has received the required boost to tackle more-complex samples with recent advances in cross-linking chemistry with MS-cleavable or reporter-based cross-linkers and faster, more sensitive and more versatile MS platforms. This protocol provides a detailed description of our optimized conditions for a full-proteome native protein preparation followed by cross-linking using the gas-phase cleavable cross-linking reagent disuccinimidyl sulfoxide (DSSO). Following cross-linking, we demonstrate extensive sample fractionation and substantially simplified data analysis with XlinkX in Proteome Discoverer, as well as subsequent protein structure investigations with DisVis and HADDOCK. This protocol produces data of high confidence and can be performed within ~10 d, including structural investigations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data are publicly available through PRIDE repository PXD008418.
References
Holzer, S. et al. Crystal structure of the N-terminal domain of human Timeless and its interaction with Tipin. Nucleic Acids Res. 45, 5555–5563 (2017).
Komolov, K. E. et al. Structural and functional analysis of a β2-adrenergic receptor complex with GRK5. Cell 169, 407–421 (2017).
Kim, S. J. et al. Molecular architecture of the Nup82 complex, the cytoplasmic mRNA export platform in the nuclear pore complex. Cell 167, 1215–1228 (2016).
Plaschka, C. et al. Architecture of the RNA polymerase II–mediator core initiation complex. Nature 518, 376–380 (2015).
Snijder, J. et al. Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science 355, 1181–1184 (2017).
Schilling, B., Row, R. H., Gibson, B. W., Guo, X. & Young, M. M. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 (2003).
Arlt, C., Ihling, C. H. & Sinz, A. Structure of full-length p53 tumor suppressor probed by chemical cross-linking and mass spectrometry. Proteomics 15, 2746–2755 (2015).
Legal, T., Zou, J., Sochaj, A., Rappsilber, J. & Welburn, J. P. I. Molecular architecture of the Dam1 complex – microtubule interaction. Open Biol. 6, 150327–150336 (2016).
Mosalaganti, S. et al. Structure of the RZZ complex and molecular basis of its interaction with Spindly. J. Cell Biol. 216, 961–981 (2017).
Ciferri, C. et al. Molecular architecture of human polycomb repressive complex 2. Elife 1, e00005 (2012).
Gupta, K., Watson, A. A., Baptista, T., Scheer, E. & Anna, L. Architecture of TAF11/TAF13/TBP complex suggests novel regulatory state in general transcription factor TFIID function. eLife 6, e30395 (2017).
Scott, H. et al. Spatial organization and molecular interactions of the Schizosaccharomyces pombe Ccq1–Tpz1–Poz1 Shelterin Complex. J. Mol. Biol. 429, 2863–2872 (2017).
Stucchi, R. et al. Regulation of KIF1A-driven dense core vesicle transport: Ca2+/CaM controls DCV binding and Liprin-α/TANC2 recruits DCVs to postsynaptic sites. Cell Reports 24, 685–700 (2018).
Liu, F., Lössl, P., Rabbitts, B. M., Balaban, R. S. & Heck, A. J. R. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for co-existing respiratory supercomplexes. Mol. Cell. Proteomics 16, RA117.000470 (2017).
Schweppe, D. K. et al. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl Acad. Sci. USA 114, 1732–1737 (2017).
Fasci, D. et al. Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell. Proteomics 17, 2018–2033 (2018).
Götze, M. et al. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectrom. 26, 83–97 (2014).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Kao, A. et al. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212 (2011).
Müller, M. Q., Dreiocker, F., Ihling, C. H., Schäfer, M. & Sinz, A. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 82, 6958–6968 (2010).
Hage, C., Iacobucci, C., Rehkamp, A., Arlt, C. & Sinz, A. The first ‘zero-length’ mass spectrometry-cleavable cross-linker for protein structure analysis. Angew. Chem. Int. Ed. Engl. 56, 14551–14555 (2017).
Weisbrod, C. R. et al. In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579 (2013).
De Jong, L. et al. In-culture cross-linking of bacterial cells reveals large-scale dynamic protein-protein interactions at the peptide level. J. Proteome Res. 16, 2457–2471 (2017).
Wang, X. et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 292, 16310–16320 (2017).
Liu, F., Lössl, P., Scheltema, R., Viner, R. & Heck, A. J. R. Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 8, 15473.1–15473.8 (2017).
Benda, C. et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell 56, 43–54 (2014).
Fagerlund, R. D. et al. Spacer capture and integration by a type I-F Cas1–Cas2-3 CRISPR adaptation complex. Proc. Natl Acad. Sci. USA 114, 5122–5128 (2017).
Liu, F., Rijkers, D. T. S., Post, H. & Heck, A. J. R. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 (2015).
Leitner, A. et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126 (2012).
Combe, C. W., Fischer, L. & Rappsilber, J. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteomics 14, 1137–1147 (2015).
Cline, S. M. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Pettersen, E. F. et al. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8 https://sourceforge.net/projects/pymol/files/pymol/1.8/ (2015).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40.1–40.8 (2008).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2014).
van Zundert, G. C. P. & Bonvin, A. M. J. J. DisVis: quantifying and visualizing accessible interaction space of distance-restrained biomolecular complexes. Bioinformatics 31, 3222–3224 (2015).
van Zundert, G. C. P. et al. The DisVis and PowerFit Web Servers: explorative and integrative modeling of biomolecular complexes. J. Mol. Biol. 429, 399–407 (2016).
Dominguez, C., Boelens, R. & Bonvin, A. M. J. J. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
van Zundert, G. C. P. et al. The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016).
Zhong, X. et al. Large-scale and targeted quantitative cross-linking MS using isotope-labeled protein interaction reporter (PIR) cross-linkers. J. Proteome Res. 16, 720–727 (2016).
Navare, A. T. et al. Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure 23, 762–773 (2015).
Tang, X., Munske, G. R., Siems, W. F. & Bruce, J. E. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 (2005).
Burke, A. M. et al. Synthesis of two new enrichable and MS-cleavable cross-linkers to define protein–protein interactions by mass spectrometry. Org. Biomol. Chem. 13, 5030–5037 (2015).
Tan, D. et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. eLife 5, 1–52 (2016).
Gutierrez, C. B. et al. Developing a novel acidic residue reactive and sulfoxide-containing MS-cleavable homobifunctional cross-linker for probing protein-protein interactions. Anal. Chem. 88, 8315–8322 (2016).
Wang, X. et al. Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 16, 840–854 (2017).
Herzog, F. et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 (2012).
Chavez, J. D. et al. Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 1–6 (2017).
Nowak, D. E., Tian, B. & Brasier, A. R. Two-step cross-linking method for identification of NF-kB gene network by chromatin immunoprecipitation. Biotechniques 39, 715–724 (2005).
Yu, C. et al. Characterization of dynamic UbR-proteasome subcomplexes by in vivo cross-linking (X) assisted bimolecular tandem affinity purification (XBAP) and label-free quantitation. Mol. Cell. Proteomics 15, 2279–2292 (2016).
Walker-Gray, R., Stengel, F. & Gold, M. G. Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and novel crosslinking approaches. Proc. Natl Acad. Sci. USA 114, 10414–10419 (2017).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014).
Walzthoeni, T. et al. xTract: software for characterizing conformational changes of protein complexes by quantitative cross-linking mass spectrometry. Nat. Methods 12, 1185–1190 (2015).
Fischer, L., Chen, Z. A. & Rappsilber, J. Quantitative cross-linking/mass spectrometry using isotope-labelled cross-linkers. J. Proteomics 88, 120–128 (2013).
Chen, Z. A., Fischer, L., Cox, J. & Rappsilber, J. Quantitative cross-linking/mass spectrometry using isotope-labeled cross-linkers and MaxQuant. Mol. Cell. Proteomics 15, 2769–2778 (2016).
Schmidt, C. et al. Comparative cross-linking and mass spectrometry of an intact F-type ATPase suggest a role for phosphorylation. Nat. Commun. 4, 1–11 (2013).
Chavez, J. D. et al. Quantitative interactome analysis reveals a chemoresistant edgotype. Nat. Commun. 6, 7928–7940 (2015).
Yu, C. et al. Developing a multiplexed quantitative cross-linking mass spectrometry platform for comparative structural analysis of protein complexes. Anal. Chem. 88, 10301–10308 (2016).
Rampler, E. et al. Comprehensive cross-linking mass spectrometry reveals parallel orientation and flexible conformations of plant HOP2-MND1. J. Proteome Res. 14, 5048–5062 (2015).
Kahraman, A. et al. Cross-link guided molecular modeling with ROSETTA. PLoS ONE 8, e73411 (2013).
Giansanti, P., Tsiatsiani, L., Low, T. Y. & Heck, A. J. R. Six alternative proteases for mass spectrometry–based proteomics beyond trypsin. Nat. Protoc. 11, 993–1006 (2016).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Jones, L. J., Haugland, R. P. & Singer, V. L. Development and characterization of the NanoOrange protein quantitation assay: a fluorescence-based assay of proteins in solution. Biotechniques 34, 850–861 (2003).
Hennrich, M. L., Groenewold, V., Kops, G. J. P. L., Heck, A. J. R. & Mohammed, S. Improving depth in phosphoproteomics by using a strong cation exchange-weak anion exchange-reversed phase multidimensional separation approach. Anal. Chem. 83, 7137–7143 (2011).
Ferber, M. et al. Automated structure modeling of large protein assemblies using crosslinks as distance restraints. Nat. Methods 13, 515–523 (2016).
Meiring, H. D., van der Heeft, E., ten Hove, G. J. & de Jong, A. Nanoscale LC-MS(n): technical design and applications to peptide and protein analysis. J. Sep. Sci. 25, 557–568 (2002).
Villén, J. & Gygi, S. P. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 (2008).
Udeshi, N. D., Mertins, P., Svinkina, T. & Carr, S. A. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 (2013).
Wiśniewski, J. R., Hein, M. Y., Cox, J. & Mann, M. A ‘Proteomic Ruler’ for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics 13, 3497–3506 (2014).
Koukourakis, M. I. et al. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer 89, 877–885 (2003).
Zimmerman, H. J., Pincus, M. R. & Henry, J. B. in Clinical Diagnosis and Management by Laboratory Methods (ed. Henry, J. B.) 281–284 (WB Saunders, Philadelphia, 1996).
We thank all Heck-group members for their helpful contributions, and, in particular, F. Liu for her initial development in proteome-wide cross-linking, M. Damen and D. Hagemans for SCX support, and K. Dingess and K. Stecker for their help with the manuscript. From Thermo Fisher Scientific, we thank B. Delanghe, K. Fitzemeier and F. Berg for their collaboration on incorporating the XLinkX cross-link search engine into the Proteome Discoverer software and R. Viner for her collaborative work on DSSO cross-linking and support in mass spectrometry method development. We acknowledge financial support from the large-scale proteomics facility Proteins@Work (Project 184.032.201), embedded in the Netherlands Proteomics Centre and supported by the Netherlands Organization for Scientific Research (NWO). Additional support came from the European Union Horizon 2020 program FET-OPEN project MSmed, Project 686547.
Author information
Authors and Affiliations
Contributions
O.K., A.J.R.H. and R.A.S. conceived the study. O.K. prepared the cell extracts and acquired the MS data. O.K. and R.A.S. analyzed the data. B.S., S.P. and D.F. provided various optimizations of the protocol. All authors critically read and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Fagerlund, R. D. et al. Proc. Natl Acad. Sci. USA 114, E5122–E5128 (2017): http://www.pnas.org/content/114/26/E5122.long
Liu, F. et al. Nat. Commun. 8, 15473 (2017): https://www.nature.com/articles/ncomms15473
Benda, C. et al. Mol. Cell 56, 43–54 (2014): https://www.sciencedirect.com/science/article/pii/S1097276514007096?via%3Dihub
Fasci, D., van Ingen, H., Scheltema, R. A. & Heck, J. R. Mol. Cell Proteomics 17, 2018–2033 (2018). http://www.mcponline.org/content/17/2/216.long
Acknowledgements
Integrated supplementary information
Supplementary Figure 1 Setup of crosslinker properties in Proteome Discoverer.
Setting up DSSO as chemical modification. For calculating of the final modification mass, the NHS-esters must be subtracted from the initial mass of DSSO molecule. All possible reactive sites are exemplified. According to the DSSO fragmentation pathway, different fragments are formed: Alkene, Sulfenic acid and Thiol. Monolinks also must be set here, as shown for DSSO hydrolyzed and DSSO quenched with Tris. As connected fragments characteristic for DSSO, “Alkene” to “Sulfenic” acid and “Alkene” to “Thiol” are set.
Supplementary Figure 2 DisVis interaction prediction interface between LDH A and LDH B with defined distance restraint.
(a) Left view of LDH B with possible interaction interface of LDH A. (b) Right view. (c) Top view. (d) Bottom view.
Supplementary information
Supplementary Figures 1 and 2
Supplementary Figures 1-2, Supplementary Table 1 and Supplementary Tutorial
Rights and permissions
About this article
Cite this article
Klykov, O., Steigenberger, B., Pektaş, S. et al. Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat Protoc 13, 2964–2990 (2018). https://doi.org/10.1038/s41596-018-0074-x
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0074-x
This article is cited by
-
The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons
Nature (2024)
-
Noncanonical assembly, neddylation and chimeric cullin–RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex
Nature Structural & Molecular Biology (2024)
-
Talin and kindlin use integrin tail allostery and direct binding to activate integrins
Nature Structural & Molecular Biology (2023)
-
Towards a structurally resolved human protein interaction network
Nature Structural & Molecular Biology (2023)
-
Structural surfaceomics reveals an AML-specific conformation of integrin β2 as a CAR T cellular therapy target
Nature Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.