Abstract
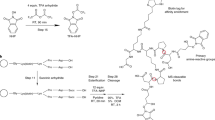
Chemical cross-linking in combination with mass spectrometric analysis of the created cross-linked products is an emerging technology aimed at deriving valuable structural information from proteins and protein complexes. The goal of our protocol is to obtain distance constraints for structure determination of proteins and to investigate protein–protein interactions. We present an integrated workflow for cross-linking/mass spectrometry (MS) based on protein cross-linking with MS-cleavable reagents, followed by enzymatic digestion, enrichment of cross-linked peptides by strong cation-exchange chromatography (SCX), and LC/MS/MS analysis. To exploit the full potential of MS-cleavable cross-linkers, we developed an updated version of the freely available MeroX software for automated data analysis. The commercially available, MS-cleavable cross-linkers (DSBU and CDI) used herein possess different lengths and react with amine as well as hydroxy groups. Owing to the formation of two characteristic 26-u doublets in their MS/MS spectra, many fewer false positives are found than when using classic, non-cleavable cross-linkers. The protocol, exemplified herein for BSA and the whole Escherichia coli ribosome, is robust and widely applicable, and it allows facile identification of cross-links for deriving spatial constraints from purified proteins and protein complexes. The cross-linking/MS procedure takes 2–3 days to complete.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
MS data and MeroX settings files are provided in the Supplementary Information.
References
Young, M. M. et al. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. USA 97, 5802–5806 (2000).
Sinz, A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 (2006).
Leitner, A. et al. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 (2010).
Petrotchenko, E. V. & Borchers, C. H. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 29, 862–876 (2010).
Rappsilber, J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 (2011).
Leitner, A., Faini, M., Stengel, F. & Aebersold, R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016).
Greber, B. J. et al. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 (2015).
Benda, C. et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell 56, 43–54 (2014).
Schmidt, C. & Urlaub, H. Combining cryo-electron microscopy (cryo-EM) and cross-linking mass spectrometry (CX-MS) for structural elucidation of large protein assemblies. Curr. Opin. Struct. Biol. 46, 157 (2017).
Chavez, J. D., Schweppe, D. K., Eng, J. K. & Bruce, J. E. In vivo conformational dynamics of Hsp90 and its interactors. Cell Chem. Biol. 23, 716–726 (2016).
Tang, X., Munske, G. R., Siems, W. F. & Bruce, J. E. Mass spectrometry identifiable cross-linking strategy for studying protein−protein interactions. Anal. Chem. 77, 311–318 (2005).
Chavez, J. D. et al. Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 136-141.e5 (2017).
Zhang, H. et al. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell. Proteomics 8, 409–420 (2009).
Zheng, C. et al. Cross-linking measurements of in vivo protein complex topologies. Mol. Cell. Proteomics 10, M110.006841 (2011).
Yang, Y. et al. Genetically encoded releasable photo-cross-linking strategies for studying protein–protein interactions in living cells. Nat. Protoc. 12, 2147–2168 (2017).
Chavez, J. D., Weisbrod, C. R., Zheng, C., Eng, J. K. & Bruce, J. E. Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteomics 12, 1451–1467 (2013).
Weisbrod, C. R. et al. In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579 (2013).
Schweppe, D. K. et al. Host-microbe protein interactions during bacterial infection. Chem. Biol. 22, 1521–1530 (2015).
Kleiner, R. E., Hang, L. E., Molloy, K. R., Chait, B. T. & Kapoor, T. M. A chemical proteomics approach to reveal direct protein-protein interactions in living cells. Cell Chem. Biol. 25, 110-120.e3 (2018).
Wu, X. et al. In vivo protein interaction network analysis reveals porin-localized antibiotic inactivation in Acinetobacter baumannii strain AB5075. Nat. Commun. 7, 13414 (2016).
Yu, C. et al. Characterization of dynamic UbR-proteasome subcomplexes by in vivo cross-linking (X) assisted bimolecular tandem affinity purification (XBAP) and label-free quantitation. Mol. Cell. Proteomics 15, 2279–2292 (2016).
Zhu, X. et al. In planta chemical cross‐linking and mass spectrometry analysis of protein structure and interaction in Arabidopsis. Proteomics 16, 1915–1927 (2016).
de Jong, L. et al. In culture cross-linking of bacterial cells reveals large scale dynamic protein-protein interactions at the peptide level. J. Proteome Res. 16, 2457-2471 (2017).
Wang, X. et al. Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 16, 840–854 (2017).
Leitner, A., Walzthoeni, T. & Aebersold, R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc. 9, 120–137 (2014).
Sinz, A. The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks. Expert Rev. Proteomics 11, 733–743 (2014).
Rinner, O. et al. Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 (2008).
Hoopmann, M. R. et al. Kojak: efficient analysis of chemically cross-linked protein complexes. J. Proteome Res. 14, 2190–2198 (2015).
Götze, M. et al. StavroX—a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectr. 23, 76–87 (2012).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Liu, F., Rijkers, D. T., Post, H. & Heck, A. J. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 (2015).
Götze, M. et al. Automated assignment ofMS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectr. 26, 83–97 (2015).
Boelt, S. G. et al. Mapping the Ca2+ induced structural change in calreticulin. J. Proteomics 142, 138–148 (2016).
Holding, A. N. XL-MS: protein cross-linking coupled with mass spectrometry. Methods 89, 54–63 (2015).
Schilling, B., Row, R. H., Gibson, B. W., Guo, X. & Young, M. M. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectr. 14, 834–850 (2003).
Teimer, R., Kosinski, J., von Appen, A., Beck, M. & Hurt, E. A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat Commun. 8, 1107 (2017).
Fernandez-Martinez, J. et al. Structure and function of the nuclear pore complex cytoplasmic mRNA export platform. Cell 167, 1215–1228.e25 (2016).
Bui, K. H. et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243 (2013).
Chen, J. et al. 6S RNA mimics B-form DNA to regulate Escherichia coli RNA polymerase. Mol. Cell 68, 388–397.e6 (2017).
Xu, Y. et al. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat. Commun. 8, 15741 (2017).
Sheppard, C. et al. Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP. Nat. Commun. 7, 13595 (2016).
Müller, M. Q. et al. A universal matrix‐assisted laser desorption/ionization cleavable cross‐linker for protein structure analysis. Rapid Commun. Mass Spectrom. 25, 155–161 (2011).
Müller, M. Q., Dreiocker, F., Ihling, C. H., Schäfer, M. & Sinz, A. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 82, 6958–6968 (2010).
Hage, C., Iacobucci, C., Rehkamp, A., Arlt, C. & Sinz, A. The first zero-length mass spectrometry-cleavable cross-linker for protein structure analysis. Angew. Chem. Int. Ed. Engl. 56, 14551–14555 (2017).
Arlt, C. et al. Integrated workflow for structural proteomics studies based on cross-linking/mass spectrometry with an MS/MS cleavable cross-linker. Anal. Chem. 88, 7930–7937 (2016).
Arlt, C. et al. An integrated mass spectrometry based approach to probe the structure of the full-length wild-type tetrameric p53 tumor suppressor. Angew. Chem. Int. Ed. Engl. 56, 275–279 (2017).
Gotze, M. et al. Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA 23, 1552–1568 (2017).
Politis, A. et al. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat. Methods 11, 403–406 (2014).
Kalkhof, S. et al. Computational modeling of laminin N‐terminal domains using sparse distance constraints from disulfide bonds and chemical cross‐linking. Proteins 78, 3409–3427 (2010).
Schweppe, D. K., Chavez, J. D. & Bruce, J. E. XLmap: an R package to visualize and score protein structure models based on sites of protein cross-linking. Bioinformatics 32, 306–308 (2015).
Brodie, N. I., Popov, K. I., Petrotchenko, E. V., Dokholyan, N. V. & Borchers, C. H. Solving protein structures using short-distance cross-linking constraints as a guide for discrete molecular dynamics simulations. Sci. Adv. 3, e1700479 (2017).
Barandun, J. et al. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 24, 944-953 (2017).
Sinz, A. Divide and conquer: cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal. Bioanal. Chem. 409, 33–44 (2017).
Back, J. et al. A new crosslinker for mass spectrometric analysis of the quaternary structure of protein complexes. J. Am. Soc. Mass Spectr. 12, 222–227 (2001).
Liu, F., Lossl, P., Scheltema, R., Viner, R. & Heck, A. J. R. Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 8, 15473 (2017).
Zhong, X. et al. Large-scale and targeted quantitative cross-linking MS using isotope-labeled protein interaction reporter (PIR) cross-linkers. J. Proteome Res. 16, 720–727 (2016).
Kao, A. H. et al. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212 (2011).
Iacobucci, C. et al. Carboxyl-photo-reactive MS-cleavable cross-linkers: unveiling a hidden aspect of diazirine-based reagents. Anal. Chem. 90, 2805–2809 (2018).
Bomgarden, R. et al. Optimization of crosslinked peptide analysis on an Orbitrap Fusion Lumos mass spectrometer. Poster note. http://tools.thermofisher.com/content/sfs/posters/PN-64854-LC-MS-Crosslinked-Peptide-IMSC2016-PN64854-EN.pdf (2017).
Bruce, J. E. In vivo protein complex topologies: sights through a cross‐linking lens. Proteomics 12, 1565–1575 (2012).
Chavez, J. D. et al. Quantitative interactome analysis reveals a chemoresistant edgotype. Nat. Commun. 6, 7928 (2015).
Petrotchenko, E. V., Serpa, J. J. & Borchers, C. H. An isotopically coded CID-cleavable biotinylated cross-linker for structural proteomics. Mol. Cell. Proteomics 10, M110.001420 (2011).
Kaake, R. M. et al. A new in vivo cross-linking mass spectrometry platform to define protein–protein interactions in living cells. Mol. Cell. Proteomics 13, 3533–3543 (2014).
Joerger, A. C. & Fersht, A. R. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 85, 375-404 (2016).
Tidow, H. et al. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc. Natl. Acad. Sci. USA 104, 12324–12329 (2007).
Okorokov, A. L. et al. The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J. 25, 5191–5200 (2006).
Bräuning, B. et al. Structure and mechanism of the two-component α-helical pore-forming toxin YaxAB. Nat. Commun. 9, 1806 (2018).
Yu, C. & Huang, L. Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 (2018).
Soderblom, E. J. & Goshe, M. B. Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78, 8059–8068 (2006).
Soderblom, E. J., Bobay, B. G., Cavanagh, J. & Goshe, M. B. Tandem mass spectrometry acquisition approaches to enhance identification of protein‐protein interactions using low‐energy collision‐induced dissociative chemical crosslinking reagents. Rapid Commun. Mass Spectrom. 21, 3395–3408 (2007).
Mak, M., Mezö, G., Skribanek, Z. & Hudecz, F. Stability of Asp-Pro bond under high and low energy collision induced dissociation conditions in the immunodominant epitope region of herpes simplex virion glycoprotein D. Rapid Commun. Mass Spectrom. 12, 837–842 (1998).
Dreiocker, F., Müller, M. Q., Sinz, A. & Schäfer, M. Collision‐induced dissociative chemical cross‐linking reagent for protein structure characterization: applied Edman chemistry in the gas phase. J. Mass Spectrom. 45, 178–189 (2010).
Müller, M. Q., Dreiocker, F., Ihling, C. H., Schäfer, M. & Sinz, A. Fragmentation behavior of a thiourea‐based reagent for protein structure analysis by collision‐induced dissociative chemical cross‐linking. J. Mass Spectrom. 45, 880–891 (2010).
Lu, Y., Tanasova, M., Borhan, B. & Reid, G. E. Ionic reagent for controlling the gas-phase fragmentation reactions of cross-linked peptides. Anal. Chem. 80, 9279–9287 (2008).
Kao, A. et al. Mapping the structural topology of the yeast 19S proteasomal regulatory particle using chemical cross-linking and probabilistic modeling. Mol. Cell. Proteomics 11, 1566–1577 (2012).
Merkley, E. D. et al. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine–lysine distances. Protein Sci. 23, 747–759 (2014).
Leitner, A. et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126 (2012).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Fritzsche, R., Ihling, C. H., Götze, M. & Sinz, A. Optimizing the enrichment of cross‐linked products for mass spectrometric protein analysis. Rapid Commun. Mass Spectrom. 26, 653–658 (2012).
Schmidt, R. & Sinz, A. Improved single-step enrichment methods of cross-linked products for protein structure analysis and protein interaction mapping. Anal. Bioanal. Chem. 409, 2393–2400 (2017).
Iacobucci, C. & Sinz, A. To be or not to be? Five guidelines to avoid misassignments in cross-linking/mass spectrometry. Anal. Chem. 89, 7832–7835 (2017).
Sinz, A. Cross‐linking/mass spectrometry for studying protein structures and protein–protein interactions: where are we now and where should we go from here? Angew. Chem. Int. Ed. Engl. 57, 6390–6396 (2018).
Brower, E. T. et al. The molecular architecture of p85α as determined by SAXS and chemical cross-linking. Cancer Res. 73, 2225 (2013).
Bomgarden, R. et al. Optimization of crosslinked peptide analysis on an Orbitrap Fusion Lumos mass spectrometer. Poster note. http://tools.thermofisher.com/content/sfs/posters/PN-64854-LC-MS-Crosslinked-Peptide-IMSC2016-PN64854-EN.pdf (2017).
Iacobucci, C., Hage, C., Schäfer, M. & Sinz, A. A novel MS-cleavable azo cross-linker for peptide structure analysis by free radical initiated peptide sequencing (FRIPS). J. Am. Soc. Mass Spectrom. 28, 2039–2053 (2017).
Iacobucci, C., Piotrowski, C., Rehkamp, A., Ihling, C. H. & Sinz, A. The first MS-cleavable, photo-thiol-reactive cross-linker for protein structural studies. J. Am. Soc. Mass Spectrom. https://doi.org/10.1007/s13361-018-1952-8 (2018).
Iacobucci, C., Schäfer, M. & Sinz, A. Free radical–initiated peptide sequencing (FRIPS)‐based cross‐linkers for improved peptide and protein structure analysis. Mass Spectrom. Rev. https://doi.org/10.1002/mas.21568 (2018).
Grifnée, E. et al. Structural characterization of protein using an enzymatic reactor. Poster. http://hdl.handle.net/2268/174922 (2014).
Olsen, J. V. et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 (2007).
Acknowledgements
A.S. acknowledges financial support by the DFG (project Si 867/15-2). C.I. was funded by the Alexander von Humboldt Foundation. M.G. was funded by the DFG (FOR855, ‘Cytoplasmic regulation of gene expression’, and GRK1591, ‘Posttranscriptional control of gene expression—mechanisms and role in pathogenesis’). We thank X. Wang and D. Tänzler for excellent technical support.
Author information
Authors and Affiliations
Contributions
C.I., M.G., and A.S. wrote the paper. C.H.I., C.P., C.A., M.S., C.H., and R.S. contributed to the Materials and Procedure sections.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Iacobucci, C. & Sinz, A. Anal. Chem. 89, 7832–7835 (2017): https://doi.org/10.1021/acs.analchem.7b02316
Iacobucci, C. et al. Anal. Chem. 90, 2805–2809 (2018): https://pubs.acs.org/doi/abs/10.1021/acs.analchem.7b04915
Müller, M. Q., Dreiocker, F., Ihling, C. H., Schäfer, M. & Sinz, A. Anal. Chem. 82, 6958–6968 (2010): https://pubs.acs.org/doi/abs/10.1021/ac101241t
Hage, C., Iacobucci, C., Rehkamp, A., Arlt, C. & Sinz, A. Angew. Chem. Ed. Engl. 56, 14551–14555 (2017): https://doi.org/10.1002/anie.201708273
Supplementary information
Supplementary Information
Supplementary Figures 1–6 and Supplementary Tables 1–3
Supplementary Dataset 1
Cross-linking data for BSA
Supplementary Dataset 2
Cross-linking data for E. coli ribosome
Rights and permissions
About this article
Cite this article
Iacobucci, C., Götze, M., Ihling, C.H. et al. A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein–protein interactions. Nat Protoc 13, 2864–2889 (2018). https://doi.org/10.1038/s41596-018-0068-8
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0068-8
This article is cited by
-
VCF1 is a p97/VCP cofactor promoting recognition of ubiquitylated p97-UFD1-NPL4 substrates
Nature Communications (2024)
-
ECL 3.0: a sensitive peptide identification tool for cross-linking mass spectrometry data analysis
BMC Bioinformatics (2023)
-
Molecular structure of soluble vimentin tetramers
Scientific Reports (2023)
-
Phosphorylation-dependent pseudokinase domain dimerization drives full-length MLKL oligomerization
Nature Communications (2023)
-
New free radical-initiated peptide sequencing (FRIPS) mass spectrometry reagent with high conjugation efficiency enabling single-step peptide sequencing
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.