Abstract
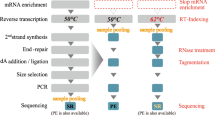
Elucidation of the complex processes involved in plant growth requires analysis of the spatial gene expression patterns in all affected tissues. This protocol extension is an adaptation of a protocol that describes how to use barcoded oligo-dT microarrays to evaluate spatial global gene expression profiles in mammalian tissue to enable it to be applied to plant material. Here, we explain the required adjustments for preparing and treating plant tissue sections on the array surface, specifically in regard to how to permeabilize and remove the tissue. Once the tissue has been removed, the cDNA–mRNA hybrid that is left on the slide is processed in the same way as cDNA obtained during experiments on mammalian tissue; thus the later stages of the protocol are not included here, and readers should follow the accompanying protocol for those. We have previously used our protocol to generate high-quality sequencing libraries for Arabidopsis thaliana inflorescence, Populus tremula developing and dormant leaf buds, and Picea abies female cones. However, we anticipate that the protocol can be adapted to other tissue types and species. The entire protocol for preparing samples and processing libraries can be completed in 3–4 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Emmert-buck, M. R. et al. Laser capture microdissection. Science 274, 998–1001 (1996).
Blokhina, O. et al. Laser capture microdissection protocol for xylem tissues of woody plants. Front. Plant Sci. 7, 1–14 (2017).
Birnbaum, K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 (2003).
Carter, A. D., Bonyadi, R. & Gifford, M. L. The use of fluorescence-activated cell sorting in studying plant development and environmental responses. Int. J. Dev. Biol. 57, 545–552 (2013).
Deal, R. B. & Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 (2010).
Moreno-romero, J., Santos-gonzález, J., Hennig, L. & Köhler, C. Applying the INTACT method to purify endosperm nuclei and to generate parental-specific epigenome profiles. Nat. Protoc. 12, 238–254 (2017).
Femino, A. M., Fay, F. S., Fogarty, K. & Singer, R. H. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Lyubimova, A. et al. Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protoc. 8, 1743–1758 (2013).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090-1–aaa6090-14 (2015).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 113, 11046–11051 (2016).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016).
Eng, C.-H. L., Shah, S., Thomassie, J. & Cai, L. Profiling the transcriptome with RNA SPOTs. Nat. Methods 14, 1153–1155 (2017).
Choi, H. M. T., Beck, V. A. & Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8, 4284–4294 (2014).
Shah, S. et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016).
Ke, R. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013).
Lee, J. H. et al. Highly multiplexed subcellular RNA sequencing in situ. Science 343, 1360–1363 (2014).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Duncan, S., Olsson, T. S. G., Hartley, M., Dean, C. & Rosa, S. A method for detecting single mRNA molecules in Arabidopsis thaliana. Plant Methods 12, 1–10 (2016).
Giacomello, S. et al. Spatially resolved transcriptome profiling in model plant species. Nat. Plants 17061, 1–11 (2017).
Lieben, L. Spatial transcriptomics in plants. Nat. Rev. Genet. 18, 394 (2017).
Salmén, F. et al. Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat. Protoc. https://doi.org/10.1038/s41596-018-0045-2 (2018).
Birnbaum, K. et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 (2005).
Deal, R. B. & Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 19, 56–68 (2010).
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 302, 801–806 (2007).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. A spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Karaiskos, N. et al. The Drosophila embryo at single-cell transcriptome resolution. Science 3235, 1–14 (2017).
Keegstra, K. Plant cell walls. Plant Physiol. 154, 483–486 (2010).
Sarkar, P., Bosneaga, E. & Auer, M. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J. Exp. Bot. 60, 3615–3635 (2009).
Burton, R. A. & Fincher, G. B. (1,3;1,4)-β-D-glucans in cell walls of the poaceae, lower plants, and fungi: a tale of two linkages. Mol. Plant 2, 873–882 (2009).
Popper, Z. A. & Fry, S. C. Primary cell wall composition of bryophytes and charophytes. Ann. Bot. 91, 1–12 (2003).
Bourgaud, F., Gravot, A., Milesi, S. & Gontier, E. Production of plant secondary metabolites: a historical perspective. Plant Sci. 161, 839–851 (2001).
Koonjul, P. K., Brandt, W. F., Farrant, J. M. & Lindsey, G. G. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res. 27, 915–916 (1999).
Yadav, R. K., Tavakkoli, M., Xie, M., Girke, T. & Reddy, G. V. A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development 141, 2735–2744 (2014).
Islam, S. et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014).
Navarro, J. F., Sjöstrand, J., Salmén, F., Lundeberg, J. & Ståhl, P. L. ST Pipeline: an automated pipeline for spatial mapping of unique transcripts. Bioinformatics 33, 2591–2593 (2017).
Acknowledgements
This work was supported by the Knut and Alice Wallenberg Foundation and the Swedish Research Council.
Author information
Authors and Affiliations
Contributions
S.G. and J.L. designed the research. S.G. developed the protocol and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.L. is the founder of a company that holds IP rights to the presented technology. S.G. declares no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Giacomello, S. et al. Nat. Plants 3, 17061 (2017): https://doi.org/10.1038/nplants.2017.61
This protocol is an extension to: Nat. Protoc. https://doi.org/10.1038/s41596-018-0045-2
Rights and permissions
About this article
Cite this article
Giacomello, S., Lundeberg, J. Preparation of plant tissue to enable Spatial Transcriptomics profiling using barcoded microarrays. Nat Protoc 13, 2425–2446 (2018). https://doi.org/10.1038/s41596-018-0046-1
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-018-0046-1
This article is cited by
-
Spatial co-transcriptomics reveals discrete stages of the arbuscular mycorrhizal symbiosis
Nature Plants (2024)
-
Plant biotechnology research with single-cell transcriptome: recent advancements and prospects
Plant Cell Reports (2024)
-
Spatial metatranscriptomics resolves host–bacteria–fungi interactomes
Nature Biotechnology (2023)
-
Understanding plant pathogen interactions using spatial and single-cell technologies
Communications Biology (2023)
-
Multiplexed single-cell 3D spatial gene expression analysis in plant tissue using PHYTOMap
Nature Plants (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.