Abstract
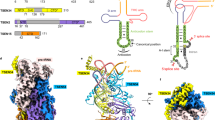
Heterotetrameric human transfer RNA (tRNA) splicing endonuclease TSEN catalyzes intron excision from precursor tRNAs (pre-tRNAs), utilizing two composite active sites. Mutations in TSEN and its associated RNA kinase CLP1 are linked to the neurodegenerative disease pontocerebellar hypoplasia (PCH). Despite the essential function of TSEN, the three-dimensional assembly of TSEN–CLP1, the mechanism of substrate recognition, and the structural consequences of disease mutations are not understood in molecular detail. Here, we present single-particle cryogenic electron microscopy reconstructions of human TSEN with intron-containing pre-tRNAs. TSEN recognizes the body of pre-tRNAs and pre-positions the 3′ splice site for cleavage by an intricate protein-RNA interaction network. TSEN subunits exhibit large unstructured regions flexibly tethering CLP1. Disease mutations localize far from the substrate-binding interface and destabilize TSEN. Our work delineates molecular principles of pre-tRNA recognition and cleavage by human TSEN and rationalizes mutations associated with PCH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-14925 (full-length TSEN–pre-tRNATyrGTA complex) and EMD-14923 (truncated TSEN–pre-tRNAArgTCT complex). Atomic coordinates of truncated TSEN–pre-tRNAArgTCT complex were deposited to the Protein Data Bank (http://www.rcsb.org) under accession number 7ZRZ. MD simulation datasets were uploaded to the Zenodo Open Science repository (https://doi.org/10.5281/zenodo.6513519). Reference models were derived from the AlphaFold Protein Structure Database with identifiers AF-Q8NCE0-F1, AF-Q8WW01-F1, AF-Q9BSV6-F1, and AF-Q7Z6J9-F1, or retrieved from the protein data bank (PDB) with accession codes 6GJW, 6Z9U, 3L0U, and 1EHZ. All data are available in the main text, public repositories, or the supplementary materials. Source data are provided with this paper.
Code availability
All codes used in this study are freely available to academic users via the indicated resources.
References
Schimmel, P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19, 45–58 (2018).
Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016).
Paushkin, S. V., Patel, M., Furia, B. S., Peltz, S. W. & Trotta, C. R. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117, 311–321 (2004).
Popow, J. et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 (2011).
Rauhut, R., Green, P. R. & Abelson, J. Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J. Biol. Chem. 265, 18180–18184 (1990).
Song, J. & Markley, J. L. Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold. J. Mol. Biol. 366, 155–164 (2007).
Sekulovski, S. et al. Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia. Nat. Commun. 12, 5610 (2021).
Tocchini-Valentini, G. D., Fruscoloni, P. & Tocchini-Valentini, G. P. Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc. Natl Acad. Sci. USA 102, 8933–8938 (2005).
Hirata, A. et al. X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity. Nucleic Acids Res. 40, 10554–10566 (2012).
Trotta, C. R. et al. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858 (1997).
Li, H., Trotta, C. R. & Abelson, J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280, 279–284 (1998).
Hayne, C. K., Schmidt, C. A., Haque, M. I., Matera, A. G. & Stanley, R. E. Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage. Nucleic Acids Res. 48, 7609–762 (2020).
Lee, M. C. & Knapp, G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J. Biol. Chem. 260, 3108–3115 (1985).
Swerdlow, H. & Guthrie, C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J. Biol. Chem. 259, 5197–5207 (1984).
Thompson, L. D. & Daniels, C. J. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J. Biol. Chem. 265, 18104–18111 (1990).
Reyes, V. M. & Abelson, J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55, 719–730 (1988).
Greer, C. L., Söll, D. & Willis, I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 76–84 (1987).
Baldi, M. I., Mattoccia, E., Bufardeci, E., Fabbri, S. & Tocchini-Valentini, G. P. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 255, 1404–1408 (1992).
Di Nicola Negri, E. et al. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89, 859–866 (1997).
Schmidt, C. A., Giusto, J. D., Bao, A., Hopper, A. K. & Matera, A. G. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res. 47, 6452–6465 (2019).
Xue, S., Calvin, K. & Li, H. RNA recognition and cleavage by a splicing endonuclease. Science 312, 906–910 (2006).
Trotta, C. R., Paushkin, S. V., Patel, M., Li, H. & Peltz, S. W. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441, 375–377 (2006).
Weitzer, S. & Martinez, J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447, 222–226 (2007).
Hanada, T. et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495, 474–480 (2013).
Karaca, E. et al. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell 157, 636–650 (2014).
Schaffer, A. E. et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 157, 651–663 (2014).
Monaghan, C. E., Adamson, S. I., Kapur, M., Chuang, J. H. & Ackerman, S. L. The Clp1 R140H mutation alters tRNA metabolism and mRNA 3′ processing in mouse models of pontocerebellar hypoplasia. Proc. Natl Acad. Sci. USA 118, e2110730118 (2021).
Schmidt, C. A. et al. Mutations in Drosophila tRNA processing factors cause phenotypes similar to pontocerebellar hypoplasia. Biol. Open 11, bio058928 (2022).
van Dijk, T., Baas, F., Barth, P. G. & Poll-The, B. T. What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet J. Rare Dis. 13, 92 (2018).
Weitzer, S., Hanada, T., Penninger, J. M. & Martinez, J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley Interdiscip. Rev. RNA 6, 47–63 (2015).
Schmidt, C. A. & Matera, A. G. tRNA introns: presence, processing, and purpose. Wiley Interdiscip. Rev. RNA 11, e1583 (2020).
Gogakos, T. et al. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 20, 1463–1475 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Byrne, R. T., Konevega, A. L., Rodnina, M. V. & Antson, A. A. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 38, 4154–4162 (2010).
Bufardeci, E., Fabbri, S., Baldi, M. I., Mattoccia, E. & Tocchini-Valentini, G. P. In vitro genetic analysis of the structural features of the pre-tRNA required for determination of the 3′ splice site in the intron excision reaction. EMBO J. 12, 4697–4704 (1993).
Belfort, M. & Weiner, A. Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell 89, 1003–1006 (1997).
Budde, B. S. et al. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 40, 1113–1118 (2008).
Namavar, Y. et al. Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain 134, 143–156 (2011).
Bierhals, T., Korenke, G. C., Uyanik, G. & Kutsche, K. Pontocerebellar hypoplasia type 2 and TSEN2: review of the literature and two novel mutations. Eur. J. Med. Genet. 56, 325–330 (2013).
Alazami, A. M. et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 10, 148–161 (2015).
Breuss, M. W. et al. Autosomal-recessive mutations in the tRNA splicing endonuclease subunit TSEN15 cause pontocerebellar hypoplasia and progressive microcephaly. Am. J. Hum. Genet 99, 785 (2016).
de Vries, H. et al. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19, 5895–5904 (2000).
Noble, C. G., Beuth, B. & Taylor, I. A. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 35, 87–99 (2007).
Miao, F. & Abelson, J. Yeast tRNA-splicing endonuclease cleaves precursor tRNA in a random pathway. J. Biol. Chem. 268, 672–677 (1993).
Hurtig, J. E. et al. Comparative parallel analysis of RNA ends identifies mRNA substrates of a tRNA splicing endonuclease-initiated mRNA decay pathway. Proc. Natl Acad. Sci. USA 118, e2020429118 (2021).
Berger, I., Fitzgerald, D. J. & Richmond, T. J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 22, 1583–1587 (2004).
Li, M. Z. & Elledge, S. J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007).
Reyes, V. M. & Abelson, J. A synthetic substrate for tRNA splicing. Anal. Biochem. 166, 90–106 (1987).
Easton, L. E., Shibata, Y. & Lukavsky, P. J. Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography. RNA 16, 647–653 (2010).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Kidmose, R. T. et al. Namdinator — automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr 66, 213–221 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr 66, 12–21 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Brooks, B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Zgarbova, M. et al. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 7, 2886–2902 (2011).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Acknowledgements
We thank P. Devant, T.J. Heinke, and L. Sagert, Institute of Biochemistry, Goethe University Frankfurt, for biochemical characterization of TSEN and support in cell culture. We are grateful to T. Gewering and A. Möller for initial single-particle EM analyses and C. Kraft for technical support during cryo-EM data collection. Electron cryo-microscopy was carried out in the cryo-EM-facility of the Julius-Maximilians University Würzburg, and the cryo-EM infrastructure of the Institute of Biochemistry, Goethe University Frankfurt, and Collaborative Research Center 1507 (Z02–high-resolution cryo-EM platform). We thank S. Weitzer and J. Martinez for critical reading of the manuscript. This study was supported by the Collaborative Research Center 902 ‘Molecular Principles of RNA-Based Regulation’ and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – INST 161/977-1 FUGG. S.S. acknowledges a Boehringer Ingelheim Fonds fellowship. R.T. acknowledges funding by the European Research Council (ERC Advanced Grant No. 789121) and the German Research Foundation (Grant No. TA157/15-1). S.T. acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – TR 1711/1-1.
Author information
Authors and Affiliations
Contributions
S.S. produced and purified protein–RNA complexes, performed biochemical assays, and prepared cryo-EM grids. L.S. prepared and screened cryo-EM grids, collected cryo-EM data, and guided data processing carried out by S.S. and S.T. S.S. and S.T. built the atomic model. L.S.S. performed all-atom molecular dynamics simulations. S.S. and S.T designed the experiments and wrote the initial draft of the manuscript with input from all authors. R.T. acquired funding. S.T. conceived the project, acquired funding, and supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Anita Hopper and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Sara Osman was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Biochemical and biophysical properties of TSEN/pre-tRNA complexes.
a, Size exclusion chromatograms of TSEN/pre-tRNATyrGTA complex and single components. Migration profile on a Superdex 200 Increase 3.2/300 column of TSEN/pre-tRNATyrGTA complex (black), TSEN only (cyan) and pre-tRNATyrGTA only (grey). b, SDS-PAGE analysis of truncated TSEN (TSENcore) (n = 3; independent biological replicates). c, Endonuclease activity assay. Yeast pre-tRNAPheGAA was incubated without (-) and with TSENcore and analyzed by urea-PAGE (n = 2; independent biological replicates). d, Endonuclease activity assay with full-length TSEN and TSENcore. Pre-tRNAArgTCT or pre-tRNATyrGTA was incubated without (-) and with full-length and truncated TSEN complex as indicated and analyzed by urea-PAGE (n = 3; independent biological replicates). e, Simulated flexibility of TSEN/pre-tRNAArgTCT. A hybrid model of truncated TSEN/pre-tRNAArgTCT with the central TSEN2 domain (residues 1-143 and 217-459) from the AlphaFold structure prediction33 and in silico modeled intron bases 37 to 44 was subjected to all-atom molecular dynamics simulations for assessment of structural flexibility. Color-coding is according to per residue B factor (Å2) with flexible regions in red and rigid regions in blue. The transparent local resolution filtered map of the complex is superimposed on the model.
Extended Data Fig. 2 Cryo-EM data processing workflow of human TSEN (detailed in Methods).
a,b,e, Representative micrographs of two TSEN/pre-tRNATyrGTA datasets (a,b) (1,135 and 1,096 micrographs, respectively) and truncated TSEN/pre-tRNAArgTCT (8,611 micrographs) (e). White size markers indicate 50 nm. c,f, Workflow of cryo-EM data processing for TSEN/pre-tRNATyrGTA (c) and truncated TSEN/pre-tRNAArgTCT (f) with final map and resolution. d,g, Representative 2D class images of TSEN/pre-tRNATyrGTA (d) and TSEN/pre-tRNAArgTCT (g). White size markers indicate 10 nm. h, Heterorefined ab initio models of TSEN/pre-tRNAArgTCT. as shown in (f). Percentages indicate particle distribution among maps. The model in dotted box was further refined by non-uniform refinement.
Extended Data Fig. 3 Quality of cryo-EM maps.
a,b, Gold-standard Fourier shell correlation curves, local resolution maps, and Azimuth plots of TSEN/pre-tRNATyrGTA (a) and TSEN/pre-tRNAArgTCT (b). Resolution values are derived from corrected curves without FSC-mask auto-tightening. The same color key was used for both local resolution maps. c-g, Representative cryo-EM densities enclosing the atomic models of pre-tRNAArgTCT secondary and tertiary structure base pairs (c), TSEN subunits (d), 3’ bulge cation-π sandwich (e), the 3’ splice site (f), and an interaction of TSEN2 N413 with the G-U wobble base pair of the A-I helix (g).
Extended Data Fig. 4 Structural aspects of TSEN/pre-tRNA complexes.
a, Surface representation of TSEN54 and TSEN34 highlight their common N-terminal domains (NTD, cartoon representation). CTD – C-terminal domain, NTE – N-terminal extension. b, Superposition of E. coli tRNAPheGAA (cyan; PDI ID 3L0U) onto human pre-tRNAArgTCT (grey, this work).
Extended Data Fig. 5 Electrostatic interactions of human and A. fulgidus splicing endonuclease.
a, Electrostatic surface potential of human TSEN with pre-tRNA (grey). b, Electrostatic surface potential of Archaeoglobus fulgidus endonuclease (PDB ID 2GJW) with BHB-RNA (grey). For comparison, the Archaeal endonuclease structure was superimposed with the cryo-EM model of TSEN-pre-tRNAArgTCT.
Extended Data Fig. 6 Molecular details of TSEN interactions in regions of PCH mutation hotspots.
a, TSEN54 (green) S85 environment in proximity to pre-tRNA acceptor stem (grey). b, Environment of the TSEN54 Y119D mutation potentially impacting pre-tRNA (grey) recognition. c, Environment of the TSEN15 (yellow) W76 environment. TSEN34 is shown in slate blue. d, TSEN15 Y152 environment.
Supplementary information
Source data
Source Data Fig. 4
Unprocessed gel.
Source Data Extended Data Fig. 1a
Table for size exclusion chromatography runs.
Source Data Extended Data Fig. 1
Unprocessed gel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sekulovski, S., Sušac, L., Stelzl, L.S. et al. Structural basis of substrate recognition by human tRNA splicing endonuclease TSEN. Nat Struct Mol Biol 30, 834–840 (2023). https://doi.org/10.1038/s41594-023-00992-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-00992-y
This article is cited by
-
Captured: the elusive eukaryotic tRNA splicing enzyme
Nature Structural & Molecular Biology (2023)
-
Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex
Nature Communications (2023)