Abstract
DEAD-box ATPases are ubiquitous enzymes essential in all aspects of RNA biology. However, the limited in vitro catalytic activities described for these enzymes are at odds with their complex cellular roles, most notably in driving large-scale RNA remodeling steps during the assembly of ribonucleoproteins (RNPs). We describe cryo-EM structures of 60S ribosomal biogenesis intermediates that reveal how context-specific RNA unwinding by the DEAD-box ATPase Spb4 results in extensive, sequence-specific remodeling of rRNA secondary structure. Multiple cis and trans interactions stabilize Spb4 in a post-catalytic, high-energy intermediate that drives the organization of the three-way junction at the base of rRNA domain IV. This mechanism explains how limited strand separation by DEAD-box ATPases is leveraged to provide non-equilibrium directionality and ensure efficient and accurate RNP assembly.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The cryo-EM density maps and models have been deposited in EMDB and PDB with accession codes: EMD-24296/PDB-7R7A (E1 overall map/model), EMD-24290/PDB-7R72 (E1 Spb4 local map/model), EMD-24269/PDB-7NAC (E2 Overall map/model), EMD-24270/PDB-7NAD (E2 Spb4 local map/model), EMD-24271/PDB-7NAF (E2 Spb1 local map/model), EMD-24280/PDB-7R6K (E2 Noc2/Noc3 local map/model), EMD-24297/PDB-7R7C (E2 L1 stalk local map/model), EMD-24286/PDB-7R6Q (E2 foot local map/model), EMD-26259/PDB-7U0H (NE1 Overall map/model). Source data are provided with this paper.
References
Jankowsky, E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 36, 19–29 (2011).
Linder, P. & Jankowsky, E. From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 (2011).
Jarmoskaite, I. & Russell, R. RNA helicase proteins as chaperones and remodelers. Annu. Rev. Biochem. 83, 697–725 (2014).
Fairman-Williams, M. E., Guenther, U. P. & Jankowsky, E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 20, 313–324 (2010).
Yang, Q. & Jankowsky, E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 13, 981–986 (2006).
Gilman, B., Tijerina, P. & Russell, R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans. 45, 1313–1321 (2017).
Mallam, A. L. et al. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature 490, 121–125 (2012).
Russell, R., Jarmoskaite, I. & Lambowitz, A. M. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol. 10, 44–55 (2013).
Liu, F., Putnam, A. & Jankowsky, E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl Acad. Sci. USA 105, 20209–20214 (2008).
Bhaskaran, H. & Russell, R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature 449, 1014–1018 (2007).
Guenther, U. P. et al. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature 559, 130–134 (2018).
Martin, R., Straub, A. U., Doebele, C. & Bohnsack, M. T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 10, 4–18 (2013).
Rodriguez-Galan, O., Garcia-Gomez, J. J. & de la Cruz, J. Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives. Biochim. Biophys. Acta 1829, 775–790 (2013).
Klinge, S. & Woolford, J. L. Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 (2019).
Kressler, D., Hurt, E. & Bassler, J. A puzzle of life: crafting ribosomal subunits. Trends Biochem. Sci. 42, 640–654 (2017).
Gamalinda, M. et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 28, 198–210 (2014).
Kressler, D., de la Cruz, J., Rojo, M. & Linder, P. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 1855–1865 (1998).
Daugeron, M. C., Kressler, D. & Linder, P. Dbp9p, a putative ATP-dependent RNA helicase involved in 60S-ribosomal-subunit biogenesis, functionally interacts with Dbp6p. RNA 7, 1317–1334 (2001).
Rocak, S., Emery, B., Tanner, N. K. & Linder, P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 33, 999–1009 (2005).
Daugeron, M. C. & Linder, P. Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA 4, 566–581 (1998).
Zagulski, M., Kressler, D., Becam, A. M., Rytka, J. & Herbert, C. J. Mak5p, which is required for the maintenance of the M1 dsRNA virus, is encoded by the yeast ORF YBR142w and is involved in the biogenesis of the 60S subunit of the ribosome. Mol. Genet. Genomics 270, 216–224 (2003).
Ripmaster, T. L., Vaughn, G. P. & Woolford, J. L. Jr. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl Acad. Sci. USA 89, 11131–11135 (1992).
Burger, F., Daugeron, M. C. & Linder, P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 28, 2315–2323 (2000).
de la Cruz, J., Kressler, D., Rojo, M., Tollervey, D. & Linder, P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA 4, 1268–1281 (1998).
Houseley, J. & Tollervey, D. The many pathways of RNA degradation. Cell 136, 763–776 (2009).
Woolford, J. L. Jr. & Baserga, S. J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
McCann, K. L., Charette, J. M., Vincent, N. G. & Baserga, S. J. A protein interaction map of the LSU processome. Genes Dev. 29, 862–875 (2015).
Manikas, R. G., Thomson, E., Thoms, M. & Hurt, E. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res. 44, 1800–1812 (2016).
Bruning, L. et al. RNA helicases mediate structural transitions and compositional changes in pre-ribosomal complexes. Nat. Commun. 9, 5383 (2018).
Choudhury, P., Kretschmer, J., Hackert, P., Bohnsack, K. E. & Bohnsack, M. T. The DExD box ATPase DDX55 is recruited to domain IV of the 28S ribosomal RNA by its C-terminal region. RNA Biol. 18, 1124–1135 (2021).
Sailer, C. et al. A comprehensive landscape of 60S ribosome biogenesis factors. Cell Rep. 38, 110353 (2022).
Aquino, G. R. R. et al. The RNA helicase Dbp7 promotes domain V/VI compaction and stabilization of inter-domain interactions during early 60S assembly. Nat. Commun. 12, 6152 (2021).
Jaafar, M. et al. Association of snR190 snoRNA chaperone with early pre-60S particles is regulated by the RNA helicase Dbp7 in yeast. Nat. Commun. 12, 6153 (2021).
Sanghai, Z. A. et al. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature 556, 126–129 (2018).
Kater, L. et al. Construction of the central protuberance and L1 stalk during 60S subunit biogenesis. Mol. Cell 79, 615–628 (2020).
Kater, L. et al. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171, 1599–1610 (2017).
Bassler, J. et al. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell 38, 712–721 (2010).
Ulbrich, C. et al. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138, 911–922 (2009).
Burlacu, E. et al. High-throughput RNA structure probing reveals critical folding events during early 60S ribosome assembly in yeast. Nat. Commun. 8, 714 (2017).
Wurm, J.P., Glowacz, K.A. & Sprangers, R. Structural basis for the activation of the DEAD-box RNA helicase DbpA by the nascent ribosome. Proc. Natl Acad. Sci. USA 118, e2105961118 (2021).
Jaskolowski, M. et al. Structural insights into the mechanism of mitoribosomal large subunit biogenesis. Mol. Cell 79, 629–644 (2020).
Scott, M. S., Boisvert, F. M., McDowall, M. D., Lamond, A. I. & Barton, G. J. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 38, 7388–7399 (2010).
Wegrecki, M., Rodriguez-Galan, O., de la Cruz, J. & Bravo, J. The structure of Erb1–Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes. Nucleic Acids Res. 43, 11017–11030 (2015).
Thoms, M., Ahmed, Y. L., Maddi, K., Hurt, E. & Sinning, I. Concerted removal of the Erb1–Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res. 44, 926–939 (2016).
Le Hir, H. & Andersen, G. R. Structural insights into the exon junction complex. Curr. Opin. Struct. Biol. 18, 112–119 (2008).
Andersen, C. B. et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972 (2006).
Bono, F., Ebert, J., Lorentzen, E. & Conti, E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725 (2006).
Liu, F., Putnam, A. A. & Jankowsky, E. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry 53, 423–433 (2014).
Garcia-Gomez, J. J. et al. Dynamics of the putative RNA helicase Spb4 during ribosome assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 31, 4156–4164 (2011).
Montpetit, B. et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242 (2011).
Karbstein, K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 23, 242–250 (2013).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Petracek, M. E. & Longtine, M. S. PCR-based engineering of yeast genome. Methods Enzymol. 350, 445–469 (2002).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Bai, X. C. et al. An atomic structure of human gamma-secretase. Nature 525, 212–217 (2015).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Wu, S., Tan, D., Woolford, J. L. Jr., Dong, M. Q. & Gao, N. Atomic modeling of the ITS2 ribosome assembly subcomplex from cryo-EM together with mass spectrometry-identified protein-protein crosslinks. Protein Sci. 26, 103–112 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Barandun, J. et al. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 24, 944–953 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Acknowledgements
We thank M. Rosen, L. Rice, J. Berger and X. Bai for discussions and M. Parker for help with live-cell imaging. We thank D. Stoddard at the UTSW Cryo-Electron Microscopy Facility, funded in part by the CPRIT Core Facility Support Award RP170644, and J. Chen at the UTSW Structural Biology Lab. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research, with the assistance of T. Humphreys. F. S. was supported by funding from the German Research Foundation (STE 2517/1 and STE 2517/5-1) and its Collaborative Research Centre 969 (Project A06). J. P. E. was supported by the Cancer Prevention and Research Institute of Texas (RR150074), the Welch Foundation (I-1897), the UTSW Endowed Scholars Fund and the National Institutes of Health (GM135617-01).
Author information
Authors and Affiliations
Contributions
V. E. C., C. S. W and J. P. E. conceived the project. V. E. C., K. S., C. S. W., N. P. and S. B. constructed strains. V. E. C. and C. S. W. conducted all growth and microscopy assays. C. S. and F. S. provided key proteomics resources. V. E. C., K. S. and N. P. carried out sample preparations and collected cryo-EM data. V. E. C., K. S. and J. P. E. processed the cryo-EM data. The paper was written by J. P. E. and V. E. C. and edited by V. E. C., C. S. W. and J. P. E. with input from all authors. J. P. E. and F.S. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Ben Luisi, David Tollervey, John Woolford and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor: Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
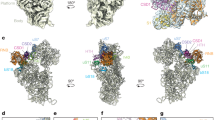
Extended Data Fig. 1 Position of the domain IV elements engaged by Spb4 within the secondary structure of the mature 25 S rRNA.
Secondary structure diagram of the mature 25S and 5.8S rRNA, showing the major subdomains with their root helices highlighted and labeled. The region in domain IV that is remodeled by Spb4 is boxed and rRNA helices described in the text are labeled and colored as in Fig. 2.
Extended Data Fig. 2 Purified samples of Ytm1∆UBL/Spb4 and of Nop53/Spb1 particles used for Cryo-EM single particle reconstruction.
Coomassie-stained 4−20% SDS−PAGE Gel of purified (a) Ytm1∆UBL/Spb4WT or (b) Nop53/Spb1 pre−ribosomal intermediates. Ytm1∆UBL/Spb4WT is was independently replicated four times while the Nop53/Spb1 was replicated twice. Protein identification was determined based on molecular weight, ambiguous bands were identified by mass spectrometry. Asterisks indicate proteins that have been tagged and therefore run higher than expected.
Extended Data Fig. 3 Micrographs, 2D classes and Fourier shell correlation (FSC) curves for reconstructions and model validation.
a, Representative electron micrograph from a total of 5122 micrographs and 2D classes of the Ytm1∆UBL/Spb4WT sample that yielded States E1 and E2. b, FSC curves for the overall and Spb4-focused maps corresponding to State E1. c, d, Local resolution filtered maps (left) and model-to-map FSC curves for each half map and full map (right) for the overall and Spb4-focused local map for State E1. e, FSC curves for the Overall, Spb4-focused and Spb1-focused map from State E2 (left). FSC curves for the peripheral maps focused on the foot, Noc2 and the L1-stalk from State E2 (right). f−k, Local resolution filtered maps (left) and model-to-map FSC curves for each half map and full map (right) for the overall and local maps corresponding to State E2. l, Representative electron micrograph from a total of 6978 micrographs and 2D classes of the Spb1/Nop53 sample that yielded State NE1. m, FSC curves for the Overall map corresponding to State NE1. n, Local resolution filtered map (left) and model-to-map FSC curves for each half map and full map (right) for the overall map corresponding to State NE1.
Extended Data Fig. 4 Data processing, classification and sorting scheme for State E1 and State E2.
Workflow of the data processing strategy after 2D classification. Map volumes and masks (red) are shown for all critical steps. Major sorting and classification criteria, particle numbers and percentages (for each 3D classification) as well as final map resolutions are indicated. Mask selection for local refinement were chosen based on boundaries with a perceived map quality drop-off was observed. Some mask boundaries were adjusted by trial and error, allowing for the best local maps to be identified for model building and refinement.
Extended Data Fig. 5 Interactions of the biogenesis factors Loc1, Noc2 and Rrp17 in state E2 and of the newly modeled helix of Nop53 in state NE1.
a, Cartoon representation of the interaction between Rrp17 (red), Spb1-MTD (teal) and H64 (blue). A plot of residue conservation scores as implemented in Jalview66 from on an alignment of 15 representative fungal Rrp17 homologs is shown below the model. Residues with an arbitrarily chosen conservation score >7 are highlighted and shown as sticks in the structural model. b, Cartoon representation of the interaction between Rrp17 (red), eL30 (green), Spb4-D1 (yellow), the Spb1 helical element (teal) and H34 (gray). Conserved residues (identified in plot) are shown as sticks (same criteria as in panel a). c, Cartoon representation of the interaction between the C-terminal helix of Rrp17 (red), Spb1-MTD (teal) and H64 (blue). Adjacent rRNA helices H90 (green) and H92 (orange) are also shown. Conserved residues (identified in plot) are shown as sticks (same criteria as in panel A). d, Detail of the position and interactions of Loc1 within the state E2 60S particle. The cartoon representation shows the interaction of Loc1 (purple) with various RBFs and rRNA elements. Model building and validation was aided by a crosslink between Loc1 and Brx1 (green connected spheres)31. e, Cartoon representation of the Noc2 (green)/Noc3(blue) interaction highlighting the conserved Noc module (orange) of Noc3 and the extended N-terminus of Noc2, which extends into the 60 S core (inset). Positions of crosslinks between Noc2 and Noc3 and Noc2 and Nip7 are shown as green connected spheres31. The interaction between Noc2/Noc3 is analogous to the interaction between Noc4 and Nop14, revealing Noc2 and Nop14 to be structural paralogs67. f, State NE1 with Nop53 colored maroon for emphasis. The newly modeled Nop53 helix is boxed and expanded in the inset. Surface view of the helix is shown for clarity.
Extended Data Fig. 6 Data processing, classification and sorting scheme for State NE1.
Workflow of the data processing strategy after 2D classification. Map volumes and masks (red) are shown for all critical steps. Major sorting and classification criteria, particle numbers and percentages (for each 3D classification) as well as final map resolutions are indicated. Masking of the L1 stalk region allowed us to classify a small subset of pre-rotation Nog2 particles, which are a contaminant in our prep as this intermediate does not contain Spb1. The final sorting was performed to select for a subset with improved density for the Spb1-AD and Spb1-CTD regions.
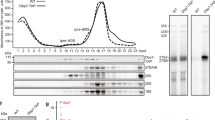
Extended Data Fig. 7 Visualization of global SHAPE analysis39 of helices H62 and H63 in early and late pre-60S particles.
a. Nucleotides in the H62/H63 region (helices are outlined and labeled) were colored according to their SHAPE reactivity values derived from experiments on early (Prp5-TAP) nucleolar pre-60S intermediates39. Nucleotides without reactivity values are shown in solid grey. b. Nucleotides in the H62/H63 region were colored according to the difference in SHAPE reactivity values between early (Prp5-TAP) and late (Nsa2-TAP) pre-60S intermediates39. The similarity in the pattern and the stability of H63 in the earliest nucleolar 60S particles indicates that H62 and H63 form a duplex structure before and after Spb4 engagement.
Extended Data Fig. 8 Structural homology to the Spb4-CTE defines a subset of DEAD-box ATPases involved in ribosome biogenesis.
a, Comparison of the CTE of Spb4, Has1 (pdb 6C0F)34, Dbp4 (Alphafold model) and Dbp7 (Alphafold model)68. Red spheres represent conserved residues among DEAD-box ATPases. b, Multiple sequence alignment of the CTE of Spb4, Has1, Dbp7 and Dbp4. The helix boundaries of Spb4 and Has1 are shown as red tubes and conserved residues, also shown in the structure cartoons, are marked with red dots. Residues within the CTD that are implicated in base specific interactions and conserved in Spb4 homologues, but not other DEAD-box proteins are indicated by green arrows and green boxes.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2
Supplementary Video 1
Animated morph between state E2 and state NE1 highlighting the rearrangement of H62 and H63 upon Spb4 release.
Source data
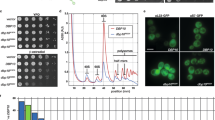
Source Data Fig. 3
Uncropped plates from spotting assays and fluorescent images.
Source Data Fig. 4
Unprocessed plates from spotting assays.
Source Data Extended Data Fig. 2
Uncropped Coomassie stained SDS–PAGE.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cruz, V.E., Sekulski, K., Peddada, N. et al. Sequence-specific remodeling of a topologically complex RNP substrate by Spb4. Nat Struct Mol Biol 29, 1228–1238 (2022). https://doi.org/10.1038/s41594-022-00874-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00874-9
This article is cited by
-
The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis
Nature Communications (2024)
-
Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles
Cell Research (2023)
-
Cellular functions of eukaryotic RNA helicases and their links to human diseases
Nature Reviews Molecular Cell Biology (2023)
-
rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly
Nature Communications (2023)