Abstract
Voltage-gated sodium channel NaV1.7 plays essential roles in pain and odor perception. NaV1.7 variants cause pain disorders. Accordingly, NaV1.7 has elicited extensive attention in developing new analgesics. Here we present cryo-EM structures of human NaV1.7/β1/β2 complexed with inhibitors XEN907, TC-N1752 and NaV1.7-IN2, explaining specific binding sites and modulation mechanism for the pore blockers. These inhibitors bind in the central cavity blocking ion permeation, but engage different parts of the cavity wall. XEN907 directly causes α- to π-helix transition of DIV-S6 helix, which tightens the fast inactivation gate. TC-N1752 induces π-helix transition of DII-S6 helix mediated by a conserved asparagine on DIII-S6, which closes the activation gate. NaV1.7-IN2 serves as a pore blocker without causing conformational change. Electrophysiological results demonstrate that XEN907 and TC-N1752 stabilize NaV1.7 in inactivated state and delay the recovery from inactivation. Our results provide structural framework for NaV1.7 modulation by pore blockers, and important implications for developing subtype-selective analgesics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The UniProt accession codes for the sequences of human Nav1.7, β1 and β2 are Q15858-3, Q07699 and O60939, respectively. The accession codes for the coordinates of Nav1.7, Nav1.5-Flecainide, Nav1.5-Propafenone and Nav1.5-Qunidine used in this study are 6J8J, 6UZ0, 7FBS and 6LQA, respectively. The accession code for the EM map of Nav1.7 used in this study is EMD-9782. The three-dimensional cryo-EM density maps of the human NaV1.7-β1-β2–XEN907, NaV1.7-β1-β2–TC-N1752 and NaV1.7-β1-β2–NaV1.7-IN2 have been deposited in the Electron Microscopy Database under accession codes EMD-33292, EMD-33295 and EMD-33296, respectively. The coordinates of the NaV1.7-β1-β2–XEN907, NaV1.7-β1-β2–TC-N1752 and NaV1.7-β1-β2–NaV1.7-IN2 have been deposited in the PDB under accession codes 7XM9, 7XMF and 7XMG, respectively. Source data are provided with this paper.
References
Catterall, W. A., Wisedchaisri, G. & Zheng, N. The chemical basis for electrical signaling. Nat. Chem. Biol. 13, 455–463 (2017).
Hille, B. Ionic Channels in Excitable Membranes 3rd edn (Oxford Univ. Press, 2001).
Catterall, W. A., Goldin, A. L. & Waxman, S. G. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57, 397–409 (2005).
Toledo-Aral, J. J. et al. Identification of PN1, a predominant voltage-dependent sodium channel expressed principally in peripheral neurons. Proc. Natl Acad. Sci. USA 94, 1527–1532 (1997).
Kanellopoulos, A. H. et al. Mapping protein interactions of sodium channel NaV1.7 using epitope-tagged gene-targeted mice. EMBO J. 37, 427–445 (2018).
Branco, T. et al. Near-perfect synaptic integration by Nav1.7 in hypothalamic neurons regulates body weight. Cell 165, 1749–1761 (2016).
Yang, Y. et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J. Med. Genet. 41, 171–174 (2004).
Dib-Hajj, S. D. et al. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 128, 1847–1854 (2005).
Fertleman, C. R. et al. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 52, 767–774 (2006).
Faber, C. G. et al. Gain of function NaV1.7 mutations in idiopathic small fiber neuropathy. Ann. Neurol. 71, 26–39 (2012).
Blesneac, I. et al. Rare NaV1.7 variants associated with painful diabetic peripheral neuropathy. Pain 159, 469–480 (2018).
Cox, J. J. et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898 (2006).
Nilsen, K. B. et al. Two novel SCN9A mutations causing insensitivity to pain. Pain 143, 155–158 (2009).
Cox, J. J. et al. Congenital insensitivity to pain: novel SCN9A missense and in-frame deletion mutations. Hum. Mutat. 31, E1670–E1686 (2010).
Weiss, J. et al. Loss-of-function mutations in sodium channel Nav1.7 cause anosmia. Nature 472, 186–190 (2011).
Bennett, D. L., Clark, A. J., Huang, J., Waxman, S. G. & Dib-Hajj, S. D. The role of voltage-gated sodium channels in pain signaling. Physiol. Rev. 99, 1079–1151 (2019).
Dib-Hajj, S. D. & Waxman, S. G. Sodium channels in human pain disorders: Genetics and pharmacogenomics. Annu. Rev. Neurosci. 42, 87–106 (2019).
Alsaloum, M., Higerd, G. P., Effraim, P. R. & Waxman, S. G. Status of peripheral sodium channel blockers for non-addictive pain treatment. Nat. Rev. Neurol. 16, 689–705 (2020).
Zakrzewska, J. M. et al. Safety and efficacy of a Nav1.7 selective sodium channel blocker in patients with trigeminal neuralgia: a double-blind, placebo-controlled, randomised withdrawal phase 2a trial. Lancet Neurol. 16, 291–300 (2017).
Cao, L. et al. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci. Transl. Med. 8, 335ra356 (2016).
Kingwell, K. Nav1.7 withholds its pain potential. Nat. Rev. Drug Discov. https://doi.org/10.1038/d41573-019-00065-0 (2019).
Deuis, J. R. et al. Analgesic effects of GpTx-1, PF-04856264 and CNV1014802 in a mouse model of NaV1.7-mediated pain. Toxins 8, 78 (2016).
Pan, X. et al. Structure of the human voltage-gated sodium channel Nav1.4 in complex with β1. Science 362, eaau2486 (2018).
Pan, X. et al. Molecular basis for pore blockade of human Na+ channel Nav1.2 by the μ-conotoxin KIIIA. Science 363, 1309–1313 (2019).
Jiang, D. et al. Structure of the cardiac sodium channel. Cell 180, 122–134.e110 (2020).
Jiang, D. et al. Open-state structure and pore gating mechanism of the cardiac sodium channel. Cell 184, 5151–5162 (2021).
Clairfeuille, T. et al. Structural basis of α-scorpion toxin action on Nav channels. Science 363, eaav8573 (2019).
Jiang, D. et al. Structural basis for voltage-sensor trapping of the cardiac sodium channel by a deathstalker scorpion toxin. Nat. Commun. 12, 128 (2021).
Li, Z. et al. Structural basis for pore blockade of the human cardiac sodium channel Nav1.5 by the antiarrhythmic drug quinidine. Angew. Chem. Int. Ed. Engl. 60, 11474–11480 (2021).
Shen, H., Liu, D., Wu, K., Lei, J. & Yan, N. Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science 363, 1303–1308 (2019).
Clatot, J. et al. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Nav1.5 α-subunits. Cardiovasc. Res. 96, 53–63 (2012).
Wang, Z. et al. Calmodulin binds to the N-terminal domain of the cardiac sodium channel NaV1.5. Channels (Austin) 14, 268–286 (2020).
Holst, A. G. et al. Sick sinus syndrome, progressive cardiac conduction disease, atrial flutter and ventricular tachycardia caused by a novel SCN5A mutation. Cardiology 115, 311–316 (2010).
Ben-Shalom, R. et al. Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol. Psychiatry 82, 224–232 (2017).
McCormack, K. et al. Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. Proc. Natl Acad. Sci. USA 110, E2724–E2732 (2013).
Ahuja, S. et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science 350, aac5464 (2015).
Chowdhury, S. et al. Discovery of XEN907, a spirooxindole blocker of NaV1.7 for the treatment of pain. Bioorg. Med. Chem. Lett. 21, 3676–3681 (2011).
Bregman, H. et al. Identification of a potent, state-dependent inhibitor of Nav1.7 with oral efficacy in the formalin model of persistent pain. J. Med. Chem. 54, 4427–4445 (2011).
Bregman, H. et al. Preparation of aryl carboxamide derivatives as sodium channel inhibitors for treatment of pain. Worldwide patent WO2011103196A1 (2011).
Cummins, T. R., Dib-Hajj, S. D. & Waxman, S. G. Electrophysiological properties of mutant Nav1.7 sodium channels in a painful inherited neuropathy. J. Neurosci. 24, 8232–8236 (2004).
Chanda, B. & Bezanilla, F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol. 120, 629–645 (2002).
Sun, J. et al. Novel SCN9A missense mutations contribute to congenital insensitivity to pain: unexpected correlation between electrophysiological characterization and clinical phenotype. Mol. Pain. 16, 1744806920923881 (2020).
Favre, I., Moczydlowski, E. & Schild, L. On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophys. J. 71, 3110–3125 (1996).
Zhao, Y. et al. Molecular basis for ligand modulation of a mammalian voltage-gated Ca2+ channel. Cell 177, 1495–1506.e1412 (2019).
Zhao, Y. et al. Cryo-EM structures of apo and antagonist-bound human CaV3.1. Nature 576, 492–497 (2019).
Ragsdale, D. S., McPhee, J. C., Scheuer, T. & Catterall, W. A. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc. Natl Acad. Sci. USA 93, 9270–9275 (1996).
Catterall, W. A., Lenaeus, M. J. & Gamal El-Din, T. M. Structure and pharmacology of voltage-gated sodium and calcium channels. Annu. Rev. Pharmacol. Toxicol. 60, 133–154 (2020).
Catterall, W. A. et al. Voltage-gated ion channels and gating modifier toxins. Toxicon 49, 124–141 (2007).
Li, X. et al. Structural basis for modulation of human NaV1.3 by clinical drug and selective antagonist. Nat. Commun. 13, 1286 (2022).
Noreng, S., Li, T. & Payandeh, J. Structural pharmacology of voltage-gated sodium channels. J. Mol. Biol. 433, 166967 (2021).
Zhang, J. et al. N-type fast inactivation of a eukaryotic voltage-gated sodium channel. Nat. Commun. 13, 2713 (2022).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank X. Huang, B. Zhu, X. Li, L. Chen and other staff members at the Center for Biological Imaging, Core Facilities for Protein Science at the Institute of Biophysics, Chinese Academy of Science (IBP, CAS) and D. Sun at the SM10 Cryo-EM Facility at the Institute of Physics, Chinese Academy of Sciences (IOP, CAS) for the support in cryo-EM data collection. We thank X.C. Zhang and Y. Zhao for their helpful discussions, X. Cui, Y. Dong, Z. Yu, Y. Wu and W. Fan for their research assistant service. This work is funded by Institute of Physics, Chinese Academy of Sciences (grant nos. E0VK101 and E2V4101 to D.J.), the National Natural Science Foundation of China (grant nos. T2221001 and 32271272 to D.J., 31871083 and 82271498 to Z.H. and 82071851 to J.G.), the Chinese Academy of Sciences Strategic Priority Research Program (grant nos. 31871083 and 81371432 to Z.H.), the Chinese National Programs for Brain Science and Brain-like intelligence technology (grant no. 2021ZD0202102 to Z.H.) and the program for the HUST Academic Frontier Youth Team (grant no. 5001170068 to J.G.).
Author information
Authors and Affiliations
Contributions
D.J. designed the experiments. J.Z., Y.L. and B.Y. prepared sample for cryo-EM study and made all the constructs. J.Z. and D.J. collected cryo-EM data. D.J. processed the data, built and refined the models. J.Z. and Y.S. prepared figures. Y.S. and Z.H. collected the electrophysiology data. J.G., Z.H. and D.J. analyzed and interpreted the results. J.Z., Y.S. and D.J. wrote the paper, and all authors reviewed and revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Vladimir Yarov-Yarovoy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Florian Ullrich, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
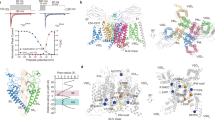
Extended Data Fig. 1 Functional characterizations and purification of NaV1.7.
a. Functional characterizations of human WT NaV1.7. Normalized conductance-voltage (G/V) relationship (Red) and steady-state fast inactivation (Blue) of NaV1.7. A schematic diagram of the recording protocol is presented below representative current traces. The Boltzmann distribution has been fitted to each data set, yielding voltage-dependence of activation V1/2 = −20.7 ± 0.7 mV (n = 13) and steady-state fast inactivation V1/2 = −76.6 ± 0.9 mV (n = 14). Data are mean + /− s.e.m. b. A representative size exclusion chromatogram profile of purified NaV1.7-β1-β2 complex. Peak fractions between green dashed lines were collected and concentrated for cryo-EM study. c. The purified NaV1.7-β1-β2 complex sample was stained with Coomassie blue on SDS-PAGE gel. NaV1.7 and β1 are labelled by red arrows. The purification was repeated independently for more than 4 times with consistent results. Source data are provided.
Extended Data Fig. 2 Cryo-EM data processing of NaV1.7-antagonist complexes.
a. The workflow of cryo-EM data processing. A total of 2,900, 2,601 and 4,430 movie stacks were collected for NaV1.7-β1-β2-XEN907 (orange), TC-N1752 (blue), and NaV1.7-IN2 (pink) respectively. Particles were auto-picked in Relion3, 2D and 3D classifications were conducted to remove bad particles, followed by 3D AutoRefine in Relion3. Subsequent Polish and CTF Refine improved image quality. According to the gold-standard Fourier Shell Correlations (FSC) criterion, the final maps were determined to 3.22, 3.09, and 3.07 Å, respectively. b-d, Sharpened map of the NaV1.7XEN (b), NaV1.7TCN (c) and NaV1.7IN2 (d) complex, colored according to the local resolution values (left). Particle angular distribution for the final 3D reconstruction (middle). FSC of the final map of the complex, calculated between two independently refined half-maps before (blue) and after (red) post-processing, overlaid with an FSC curve calculated between the cryo-EM density map and the structural model shown in black (right).
Extended Data Fig. 3 EM map of the NaV1.7XEN.
a. The EM map of the S1-S6 segment in each domain. b. The EM map for P-loop of each domain. c. The auxiliary β1 and β2 in the NaV1.7 complex are shown individually. Side- chains of residues with good density are shown in sticks. The maps were prepared in PyMOL.
Extended Data Fig. 4 The overall structure of the drug-binding complex of human NaV1.7.
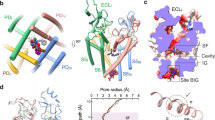
a. The EM map and model of the NaV1.7 complex viewed in parallel to the membrane plane. The β1 and β2 subunits, N-terminus domain (NTD), extracellular loops (ECLs), domain III and domain IV linker (LinkerIII-IV), and transmembrane helices S1–S6 of domain I were labeled. The α subunit was colored in cyan (DI), light red (DII), light green (DIII), light blue (DIV), and pink (LinkerIII-IV), respectively. The β1, β2 N-glycans and lipids were colored in wheat, light gray, yellow and orange, respectively. b. Overall structure comparison of human NaV1.7 α subunit-XEN907/TC-N1752/NaV1.7-IN2. The overall structures of each α subunit were colored in yellow (XEN907), blue (TC-N1752) and light pink (NaV1.7-IN2). c. Ion conductance path of NaV1.7 calculated by HOLE. The three panels on the left represent the pore radius of NaV1.7 complexed with XEN907, TC-N1752 and NaV1.7-IN2, respectively. The ligands in the cavity were omitted when calculating the pore radius. The selective filter (SF) and the intracellular activation gate (AG) were highlighted in pink and blue, respectively.
Extended Data Fig. 5 The NTD modeling and sequence alignments among human NaV channels.
a. The un-sharpened and sharpened (using DeepEMhancer) EM maps of NaV1.7XEN. b. The EM map of the N-terminus domain (NTD) was shown in surface, the secondary structure α1, α2, β1, β2 and amino acids with good density were displayed and labeled. c. AlphaFold2 model of NaV1.7 NTD. d. Structural comparison of the NaV1.7 NTD and its AlphaFold2 model. e. Gain-of-function, loss-of-function and other pathogenic mutation sites in the NTD of NaV channels are highlighted in green, red and gray, respectively. Secondary structure elements are indicated on top of the sequences. R116 in S0I of NaV1.7 mediates key interactions between the NTD and VSDI is identical among the isoforms.
Extended Data Fig. 6 Modulation of NaV1.7 by XEN907.
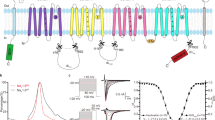
a. State-dependent inhibition of NaV1.7 by XEN907. A family of representative sodium currents in response to repetitive test pulses in the presence of 100 nM XEN907 when holding at −80 mV (Red) and −120 mV (Blue), respectively. b. Use-dependent inhibition of NaV1.7 by XEN907. Normalized current in response to repetitive test pulses in the presence of varied concentrations of XEN907 when holding at −80 mV (Red) and −120 mV (Blue), respectively. Data are mean + /− s.e.m. The n values of control, 0.01 nM, 0.1 nM, 1 nM, 10 nM, 100 nM for holding at −80 mV are 4, 3, 6, 5, 5, 4; and the n values of control, 1 nM, 10 nM, 100 nM, 1 μM, 10 μM for holding at −120 mV are 4, 7, 5, 5, 5, 4, respectively. c. The effect of 100 nM XEN907 on the voltage-dependence of activation. Data are mean + /− s.e.m. The n values for control (black) and XEN907 (red) are 6 and 8 respectively. d. The concentration-dependent shift of voltage-dependence of fast inactivation by XEN907. Data are mean + /− s.e.m. The n values of tested concentrations of control, 1 nM, 100 nM, and 10 μM are 9, 8, 12, 6, respectively. e. The EM density of XEN907. f. Pore density of NaV1.7apo (EMDB access code: EMD-9782) and NaV1.7XEN. The EM density for XEN907 is highlighted in yellow. g. EM densities S6IV of NaV1.7XEN and NaV1.7apo (PDB code: 6J8J). h. Mutants N1753M and M1754N are non-functional. Source data are provided.
Extended Data Fig. 7 Modulation of NaV1.7 by TC-N1752.
a. State-dependent inhibition of NaV1.7 by TC-N1752. A family of sodium currents in response to repetitive test pulses in the presence of 1 μM TC-N1752 when holding at −80 mV (Red) and −120 mV (Blue), respectively. b. Use-dependent inhibition of NaV1.7 by TC-N1752. Normalized current in response to repetitive test pulses in the presence of varied concentrations of TC-N1752 when holding at −80 mV (Red) and −120 mV (Blue), respectively. Data are mean + /− s.e.m. The n values of control, 0.01 μM, 0.03 μM, 0.1 μM, 0.3 μM, 1 μM, 10 μM for holding at −80 mV are 6, 4, 4, 6, 4, 4, 4; and for holding at −120 mV are 3, 4, 3, 3, 4, 5, 5, respectively. c-d. Use-dependent inhibition of NaV1.5 by TC-N1752. Data are mean + /− s.e.m. The n values of control, 1 nM, 10 nM, 0.1 μM, 1 μM, 10 μM are 3, 5, 5, 7, 10, 6, respectively. e. The dose-dependent response curve of TC-N1752 holding at −80 mV. Data are mean + /− s.e.m. The n values of 1 nM, 10 nM, 0.1 μM, 1 μM, 10 μM are 5, 4, 6, 9, 4, respectively. f. The effect of 10 μM TC-N1752 on the voltage-dependence of activation. Data are mean + /− s.e.m. The n values for control (black) and TC-N1752 (red) are 11 and 10 respectively. g. The EM density of TC-N1752. h. Pore density of NaV1.7apo (EMDB access code: EMD-9782) and NaV1.7TCN. The EM density for TC-N1752 is highlighted in blue. i. EM density for S6II of NaV1.7TCN and NaV1.7apo (PDB code: 6J8J). Source data are provided.
Extended Data Fig. 8 Modulation of NaV1.7 by NaV1.7-IN2.
a. State-independent inhibition of NaV1.7 by NaV1.7-IN2. A family of sodium currents in response to repetitive test pulses in the presence of 1 μM NaV1.7-IN2 when holding at −80 mV (Red) and −120 mV (Blue), respectively. b. Use-dependent inhibition of NaV1.7 by NaV1.7-IN2. Normalized current in response to repetitive test pulses in the presence of varied concentrations of NaV1.7-IN2 when holding at −80 mV (Red) and −120 mV (Blue), respectively. Data are mean + /− s.e.m. The n values of control, 0.1 nM, 1 nM, 10 nM, 0.1 μM, 1 μM for holding at −80 mV are 3, 3, 5, 3, 4, 3; and for holding at −120 mV are 3, 4, 3, 3, 3, 3, respectively. c, d. Use-dependent inhibition of NaV1.5 by NaV1.7-IN2. Data are mean + /− s.e.m. The n values of control, 0.1 nM, 1 nM, 10 nM, 0.1 μM, 1 μM are 4, 4, 5, 6, 4, 4, respectively. e. The dose-dependent response curve of NaV1.7-IN2 holding at −80 mV. Data are mean + /− s.e.m. The n values of 0.1 nM, 1 nM, 10 nM, 0.1 μM, 1 μM are 4, 8, 7, 4, 4, respectively. f. The effect of 100 nM NaV1.7-IN2 on the recovery from fast activation of NaV1.7. Data are mean + /− s.e.m. The n values for control (black) and NaV1.7-IN2 (red) are 8 and 7 respectively. g. The EM density of NaV1.7-IN2. h. Pore density of NaV1.7apo (EMDB access code: EMD-9782) and NaV1.7IN2. The EM density for NaV1.7-IN2 is highlighted in pink. Source data are provided.
Extended Data Fig. 9 The sequence alignments of P-loops and S6 helices of human NaV channels.
Residues that contribute to drug binding were shaded and labeled. Specifically, three NaV1.7 antagonists (XEN907, TC-N1752 and NaV1.7-IN2) and three previously reported anti-arrhythmic drugs (flecainide, PDB code: 6UZ0; propafenone, PDB code: 7FBS; quinidine, PDB code: 6LQA), were represented in different shapes and colors. The interacting residues were labeled with corresponding shapes. Residues interacting with the six blockers within 5 Å are highlighted.
Supplementary information
Supplementary Video 1
XEN907 binding induced conformational changes in NaV1.7. XEN907 binds to the central cavity of NaV1.7, the binding directly causes an α- to π-helix transition of DIV-S6 helix, which tightens the fast inactivation gate.
Supplementary Video 2
TC-N1752 binding induced conformational changes in NaV1.7. TC-N1752 binds to the central cavity of NaV1.7, the binding indirectly causes an α- to π-helix transition of DII-S6 helix, which closes the activation gate.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Uncropped gel.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Shi, Y., Huang, Z. et al. Structural basis for NaV1.7 inhibition by pore blockers. Nat Struct Mol Biol 29, 1208–1216 (2022). https://doi.org/10.1038/s41594-022-00860-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00860-1
This article is cited by
-
Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin
Nature Communications (2024)
-
Structure and electromechanical coupling of a voltage-gated Na+/H+ exchanger
Nature (2023)
-
Cannabidiol inhibits Nav channels through two distinct binding sites
Nature Communications (2023)
-
Why to Study Peptides from Venomous and Poisonous Animals?
International Journal of Peptide Research and Therapeutics (2023)