Abstract
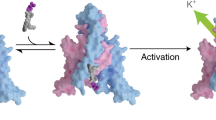
Lipids play important roles in regulating membrane protein function, but the molecular mechanisms used are elusive. Here we investigated how anionic lipids modulate SthK, a bacterial pacemaker channel homolog, and HCN2, whose activity contributes to pacemaking in the heart and brain. Using SthK allowed the reconstitution of purified channels in controlled lipid compositions for functional and structural assays that are not available for the eukaryotic channels. We identified anionic lipids bound tightly to SthK and their exact binding locations and determined that they potentiate channel activity. Cryo-EM structures in the most potentiating lipids revealed an open state and identified a nonannular lipid bound with its headgroup near an intersubunit salt bridge that clamps the intracellular channel gate shut. Breaking this conserved salt bridge abolished lipid modulation in SthK and eukaryotic HCN2 channels, indicating that anionic membrane lipids facilitate channel opening by destabilizing these interactions. Our findings underline the importance of state-dependent protein-lipid interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data supporting the findings of this paper are available from the corresponding author upon request. A reporting summary for this article is available as a Supplementary Information file. MS analysis is summarized in three supplementary data sheets and the raw data have been deposited at https://doi.org/10.6084/m9.figshare.19642617.v1. The maps for SthK in detergent and in nanodiscs have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes (SthK closed state, cAMP-bound in the presence of detergent 25981; SthK closed state, cAMP-bound in the presence of POPA 25916; SthK open-state, cAMP-bound in the presence of POPA 25917). Atomic coordinates for the three structures have been deposited in the PDB with accession codes 7TKT, 7TJ5 and 7TJ6, respectively. All other structural data used in this study for comparisons are available in EMDB (human HCN1 20846, human HCN4 0094, rabbit HCN4 12513, SthK in POPG nanodiscs 7484) and the PDB (human HCN1 5U6O, rabbit HCN4 7NP4, human HCN4 6GYN, SthK 6CJQ). Source data are provided with this paper.
References
Dickson, E. J. & Hille, B. Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J. 476, 1–23 (2019).
Hansen, S. B., Tao, X. & MacKinnon, R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 (2011).
Gao, Y., Cao, E., Julius, D. & Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016).
Qiao, P., Liu, Y., Zhang, T., Benavides, A. & Laganowsky, A. Insight into the selectivity of Kir3.2 toward phosphatidylinositides. Biochemistry 59, 2089–2099 (2020).
Hughes, T. E. T. et al. Structural insights on TRPV5 gating by endogenous modulators. Nat. Commun. 9, 4198 (2018).
She, J. et al. Structural mechanisms of phospholipid activation of the human TPC2 channel. eLife 8, e45222 (2019).
Gu, R. X. & de Groot, B. L. Lipid-protein interactions modulate the conformational equilibrium of a potassium channel. Nat. Commun. 11, 2162 (2020).
Corradi, V. et al. Lipid-Protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 4, 709–717 (2018).
Hite, R. K., Butterwick, J. A. & MacKinnon, R. Phosphatidic acid modulation of Kv channel voltage sensor function. eLife 3, e04366 (2014).
Tong, A. et al. Direct binding of phosphatidylglycerol at specific sites modulates desensitization of a ligand-gated ion channel. eLife 8, e50766 (2019).
Hénault, C. M. et al. A lipid site shapes the agonist response of a pentameric ligand-gated ion channel. Nat. Chem. Biol. 15, 1156–1164 (2019).
Clark, M. D., Contreras, G. F., Shen, R. & Perozo, E. Electromechanical coupling in the hyperpolarization-activated K. Nature 583, 145–149 (2020).
Santoro, B. & Tibbs, G. R. The HCN gene family: molecular basis of the hyperpolarization-activated pacemaker channels. Ann. N. Y. Acad. Sci. 868, 741–764 (1999).
Lee, C. H. & MacKinnon, R. Structures of the human HCN1 hyperpolarization-activated channel. Cell 168, 111–120.e11 (2017).
Zolles, G. et al. Pacemaking by HCN channels requires interaction with phosphoinositides. Neuron 52, 1027–1036 (2006).
Fogle, K. J., Lyashchenko, A. K., Turbendian, H. K. & Tibbs, G. R. HCN pacemaker channel activation is controlled by acidic lipids downstream of diacylglycerol kinase and phospholipase A2. J. Neurosci. 27, 2802–2814 (2007).
Pian, P., Bucchi, A., Robinson, R. B. & Siegelbaum, S. A. Regulation of gating and rundown of HCN hyperpolarization-activated channels by exogenous and endogenous PIP2. J. Gen. Physiol. 128, 593–604 (2006).
Saponaro, A. et al. Gating movements and ion permeation in HCN4 pacemaker channels. Mol. Cell 81, 2929–2943.e6 (2021).
Lee, C. H. & MacKinnon, R. Voltage sensor movements during hyperpolarization in the HCN channel. Cell 179, 1582–1589.e7 (2019).
Schmidpeter, P. A. M., Gao, X., Uphadyay, V., Rheinberger, J. & Nimigean, C. M. Ligand binding and activation properties of the purified bacterial cyclic nucleotide-gated channel SthK. J. Gen. Physiol. 150, 821–834 (2018).
Rheinberger, J., Gao, X., Schmidpeter, P. A. & Nimigean, C. M. Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. eLife 7, e39775 (2018).
Brams, M., Kusch, J., Spurny, R., Benndorf, K. & Ulens, C. Family of prokaryote cyclic nucleotide-modulated ion channels. Proc. Natl Acad. Sci. USA 111, 7855–7860 (2014).
Rothberg, B. S., Shin, K. S., Phale, P. S. & Yellen, G. Voltage-controlled gating at the intracellular entrance to a hyperpolarization-activated cation channel. J. Gen. Physiol. 119, 83–91 (2002).
Flynn, G. E. & Zagotta, W. N. Insights into the molecular mechanism for hyperpolarization-dependent activation of HCN channels. Proc. Natl Acad. Sci. USA 115, E8086–E8095 (2018).
Schmidpeter, P. A. M., Rheinberger, J. & Nimigean, C. M. Prolyl isomerization controls activation kinetics of a cyclic nucleotide-gated ion channel. Nat. Commun. 11, 6401 (2020).
Marchesi, A. et al. An iris diaphragm mechanism to gate a cyclic nucleotide-gated ion channel. Nat. Commun. 9, 3978 (2018).
Kesters, D. et al. Structure of the SthK carboxy-terminal region reveals a gating mechanism for cyclic nucleotide-modulated ion channels. PLoS ONE 10, e0116369 (2015).
Morgan, J. L. W., Evans, E. G. B. & Zagotta, W. N. Functional characterization and optimization of a bacterial cyclic nucleotide-gated channel. J. Biol. Chem. 294, 7503–7515 (2019).
Evans, E. G. B., Morgan, J. L. W., DiMaio, F., Zagotta, W. N. & Stoll, S. Allosteric conformational change of a cyclic nucleotide-gated ion channel revealed by DEER spectroscopy. Proc. Natl Acad. Sci. USA 117, 10839–10847 (2020).
Schmidpeter, P. A. M. & Nimigean, C. M. Correlating ion channel structure and function. Methods Enzymol. 652, 3–30 (2021).
Lee, A. G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 (2004).
Gupta, K. et al. Identifying key membrane protein lipid interactions using mass spectrometry. Nat. Protoc. 13, 1106–1120 (2018).
Barrera, N. P., Zhou, M. & Robinson, C. V. The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell Biol. 23, 1–8 (2013).
Posson, D. J., Rusinova, R., Andersen, O. S. & Nimigean, C. M. Stopped-flow fluorometric ion flux assay for ligand-gated ion channel studies. Methods Mol. Biol. 1684, 223–235 (2018).
Ingólfsson, H. I. & Andersen, O. S. Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay. Drug Dev. Technol. 8, 427–436 (2010).
Shin, J. J. & Loewen, C. J. Putting the pH into phosphatidic acid signaling. BMC Biol. 9, 85 (2011).
Latorre, R., Labarca, P. & Naranjo, D. Surface charge effects on ion conduction in ion channels. Methods Enzymol. 207, 471–501 (1992).
Bell, J. E. & Miller, C. Effects of phospholipid surface charge on ion conduction in the K+ channel of sarcoplasmic reticulum. Biophys. J. 45, 279–287 (1984).
Xue, J., Han, Y., Zeng, W., Wang, Y. & Jiang, Y. Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron 109, 1302–1313.e4 (2021).
Zheng, X. et al. Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel. Nat. Struct. Mol. Biol. 27, 625–634 (2020).
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019).
Decher, N., Chen, J. & Sanguinetti, M. C. Voltage-dependent gating of hyperpolarization-activated, cyclic nucleotide-gated pacemaker channels: molecular coupling between the S4-S5 and C-linkers. J. Biol. Chem. 279, 13859–13865 (2004).
Pian, P., Bucchi, A., Decostanzo, A., Robinson, R. B. & Siegelbaum, S. A. Modulation of cyclic nucleotide-regulated HCN channels by PIP(2) and receptors coupled to phospholipase C. Pflug. Arch. 455, 125–145 (2007).
Nava, C. et al. De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat. Genet. 46, 640–645 (2014).
Dowhan, W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66, 199–232 (1997).
Hill, W. G. et al. Isolation and characterization of the Xenopus oocyte plasma membrane: a new method for studying activity of water and solute transporters. Am. J. Physiol. Ren. Physiol. 289, F217–F224 (2005).
Robinson, C. V., Rohacs, T. & Hansen, S. B. Tools for understanding nanoscale lipid regulation of ion channels. Trends Biochem. Sci. 44, 795–806 (2019).
Neale, C., Herce, H. D., Pomès, R. & García, A. E. Can specific protein-lipid interactions stabilize an active state of the beta 2 adrenergic receptor? Biophys. J. 109, 1652–1662 (2015).
Poveda, J. A. et al. Modulation of the potassium channel KcsA by anionic phospholipids: role of arginines at the non-annular lipid binding sites. Biochim. Biophys. Acta Biomembr. 1861, 183029 (2019).
Sands, Z. A. & Sansom, M. S. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure 15, 235–244 (2007).
Tomczyk, M. M. & Dolinsky, V. W. The cardiac lipidome in models of cardiovascular disease. Metabolites 10, 254 (2020).
Pradas, I. et al. Lipidomics reveals a tissue-specific fingerprint. Front. Physiol. 9, 1165 (2018).
Suh, B. C. & Hille, B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu. Rev. Biophys. 37, 175–195 (2008).
Kasimova, M. A., Tarek, M., Shaytan, A. K., Shaitan, K. V. & Delemotte, L. Voltage-gated ion channel modulation by lipids: insights from molecular dynamics simulations. Biochim. Biophys. Acta 1838, 1322–1331 (2014).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Ritchie, T. K. et al. Chapter 11—reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Ramlaul, K., Palmer, C. M., Nakane, T. & Aylett, C. H. S. Mitigating local over-fitting during single particle reconstruction with SIDESPLITTER. J. Struct. Biol. 211, 107545 (2020).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D. Struct. Biol. 74, 814–840 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Matyash, V., Liebisch, G., Kurzchalia, T. V., Shevchenko, A. & Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008).
Hutchins, P. D., Russell, J. D. & Coon, J. J. LipiDex: an integrated software package for high-confidence lipid identification. Cell Syst. 6, 621–625.e5 (2018).
Pluskal, T., Castillo, S., Villar-Briones, A. & Oresic, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 11, 395 (2010).
Marty, M. T. et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015).
Schrecke, S. et al. Selective regulation of human TRAAK channels by biologically active phospholipids. Nat. Chem. Biol. 17, 89–95 (2021).
Acknowledgements
Cryo-EM data were produced at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (no. SF349247), NYSTAR and the National Institutes of Health (NIH) National Institute of General Medical Sciences (no. GM103310), with additional support from Agouron Institute (grant no. F00316) and NIH (grant no. OD019994), and at NYU Langone Health’s Cryo-Electron Microscopy Laboratory (RRID: SCR_019202). We thank W. Zagotta and E. Evans (University of Washington, Department of Physiology and Biophysics) for providing us with E. coli C43 cyA− cells, G. Tibbs (Weill Cornell Medicine, Department of Anesthesiology) for the pGHE vector carrying the gene for mouse HCN2 and E. Kim and S. Scheuring for critical comments on the manuscript. The work presented here was sponsored by the NIH (grant nos. GM124451 to C.M.N. and K08 GM132781 to P.M.R.) and the American Heart Association (grant no. 18POST33960309 to P.A.M.S.).
Author information
Authors and Affiliations
Contributions
P.A.M.S. and C.M.N. designed the research. P.A.M.S. performed and analyzed all stopped-flow and bilayer recordings. D.W. and H.T. performed and analyzed all MS experiments. C.V.R. supervised the MS analysis. Cryo-EM data collection and analysis was carried out by J.R. for SthK in detergent and by P.A.M.S. for SthK in nanodiscs. P.A.M.S. and P.M.R. performed and analyzed TEVC recordings. P.A.M.S. and C.M.N. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks Anna Moroni and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Florian Ullrich, in collaboration with the Nature Structural and Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM processing scheme.
(a) Cryo-EM data processing and structure validation for SthK in DDM in the presence of cAMP. (b) Cryo-EM data processing and structure validation for SthK in 3:1 DOPC:POPA nanodiscs in the presence of cAMP.
Extended Data Fig. 2 Mass spectrometry analysis of SthK.
(a) Lipidomics analysis of the phospholipids extracted from the SthK sample (top) and E. coli phospholipids (bottom). PE and PG species are annotated with the length of the acyl chains and the number of double bonds. PG is enriched in the SthK sample. (b) Identification of cardiolipins (CL) in the SthK sample. CL species are annotated with the acyl chain length and the number of double bonds (left panel). The right panel shows the MS/MS identification of CL (68:3). (c) Probability of 0, 1, 2, 3, or 4 cAMP-binding events to SthK as observed in native MS are plotted in grey, red, olive, green and blue, respectively.
Extended Data Fig. 3 Absolute quantification of co-purified lipids.
Comparison of mol % PE (blue) and PG (orange) lipids extracted from purified WT SthK (a) and SthK D226N (b) with lipids extracted from E. coli membranes (grey). Lysate refers to membranes from cells overexpressing the respective protein. Results are averaged over four repeats, error bars are s.d.
Extended Data Fig. 4 Functional assays using SthK.
(a) Quenching kinetics from a Tl+ flux assay reporting SthK channel activity in different lipid compositions (DOPC – black, 3:1 DOPC:POPG – orange, 3:1 DOPC:cardiolipin (CL) – blue) after 12 ms activation by 200 µM cAMP. Traces in the absence of cAMP are in shades of grey. (b). FSEC analysis (Superdex 200 5/150) of WT SthK extracted from liposomes as used for the stopped-flow assay (DOPC – black, 3:1 DOPC:X with X = POPE – dark blue, POPS – dark green, POPG – orange and POPA – red), excitation at 280 nm, emission at 330 nm. Signal intensity was normalized to SthK extracted from DOPC-only liposomes. (c) SDS-PAGE analysis of SthK reconstituted into different lipid compositions for the stopped-flow Tl+ flux assay is shown for WT SthK and SthK D226N. Detergent (DDM) or lipid environment for the reconstitution are indicated. DOPC and 3:1 DOPC:X were used for reconstitutions, with X = POPG (PG), POPE (PE), POPS (PS), POPA (PA), or cardiolipin (CL). Molecular weight standard used was Precision Plus Protein (Bio-Rad). (d) Comparison of the open probability (Po) values obtained from single-channel recordings (cf Fig. 2c, d). Averaged values ± s.d. (from Fig. 2d) are shown for WT SthK in the presence of POPE (dark blue), POPS (dark green), POPG (orange) and POPA (red). Mean Po values reach a stable non-zero level at negative voltages (inset). (e) Comparison of the single channel conductance values obtained from single channel recordings (cf Fig. 2c, d). Averaged values ± s.d. (from Fig. 2d) are shown for WT SthK in the presence of POPE (dark blue), POPS (dark green), POPG (orange) and POPA (red). (f) Fold-change in Po for SthK reconstituted into different lipid compositions at ±20 mV, shown relative to the value obtained in 3:1 DOPC:POPE. (g) Fold-change in single channel conductance I for SthK in different lipid compositions at ±20 mV relative to the value obtained in 3:1 DOPC:POPE. Lipid compositions for F and G are 3:1 DOPC:X, with X = POPE – dark blue, X = POPS – dark green, X = POPG – orange, X = POPA – red.
Extended Data Fig. 5 Characteristics of different lipids.
Chemical structures of the lipids used in this study are shown, their IUPAC names and simplified abbreviations are given. Net charges and phase transition temperatures (TM) are included. Information was obtained from: https://avantilipids.com.
Extended Data Fig. 6 Conformational changes during gating of SthK.
(a) One subunit of SthK in the presence of POPA and cAMP is shown (closed state: blue, open state: yellow). Models were aligned to the TMDs. (b) Backbone traces of the TM helices. (c) Backbone traces of the C-linker. (d) Backbone traces of the cyclic nucleotide-binding domain (CNBD). (e) C-linker movements during gating shown in cartoon representation (closed state: blue, open state: yellow). (f) Overlay of SthK in the closed state in the presence of POPA (blue) and POPG (grey, PDB: 6cju). (g) Alignment of only the CNBDs between closed (blue) and open (yellow) SthK. (h) Overlay of the CNBD of SthK in the open state (yellow) with the x-ray structure of the activated CNBD of SthK (purple, PDB: 4d7t).
Extended Data Fig. 7 Protein environment of the non-annular lipid along the S6 helix.
(a) Cryo-EM density of SthK in 3:1 DOPC:POPA nanodiscs with the protein in light blue, lipids in orange. Lipids that have not been investigated in this study are transparent. Zoom shows the non-annular lipid along S6 at the center of this study. (b) Non-annular lipid along S6 (orange, spheres) embedded in the protein (electrostatic surface representation calculated in Chimera X). (c) S5 and S6 helices of adjacent subunits are shown as cartoon (light and dark blue) with Arg136 on S5 and Asp226 on S6 in ball and stick representation and the lipid along the S6 helix of one subunit as density obtained from cryo-EM. Three different lipids are modelled into this density and shown in ball and stick representation (from left to right: POPA, POPG, DOPC) revealing that the PA headgroup fits best in the experimental density. (d) Cryo-EM density of SthK in 3:1 DOPC:POPA nanodiscs (grey, unsharpened map) showing continuous density between Arg136 and the exposed phosphate group of POPA. (e) Lipid (orange, ball and stick representation) along the S6 helix between two adjacent subunits (dark and light blue, cartoon, only S5 and S6 are shown for clarity). The lipid is intertwined between S5 and S6 of neighboring subunits and is in contact with the bundle crossing and the pore helix/ selectivity filter.
Extended Data Fig. 9 Conservation of the intersubunit salt-bridge in SthK and HCN channels.
(a)–(c) Cryo-EM density of HCN channels (grey) with extra-proteinaceous density shaped like lipids (red) is shown for human HCN1 (A, EMDB: 20846), human HCN4 (B, EMDB: 0094), rabbit HCN4 (C, EMBD: 12513). The top row shows an overview of the entire protein, the bottom row the zoom on the lipid along the S6 helix. (d) Overlay of one subunit of SthK (gold, PDB: 6CJQ) and HCN1 (grey, PDB: 5U6O) showing the structural homology between the bacterial and the eukaryotic proteins. (e)–(g) Adjacent subunits (light and dark grey) are shown for human HCN1 (E, PDB: 5U6O), rabbit HCN4 (F, PDB: 7NP4), human HCN4 (G, PDB: 6GYN). Overview of the entire protein is shown in the top row, the bottom row is a zoom on the inter-subunit salt-bridge for SthK and the three eukaryotic HCN channels. Cα – Cα distances between Arg on S5 and Asp on S6 are indicated for each protein to emphasize the structural conservation of the salt-bridge between the bacterial channel SthK and eukaryotic HCN channels.
Extended Data Fig. 10 Two-electrode voltage clamp recordings of mouse HCN2.
(a) TEVC recordings of oocytes expressing HCN2 after treatment with PMA (top panel) or Wortmannin (bottom panel). (b) TEVC recordings of oocytes expressing HCN2 D443N after treatment with PMA (top panel) or Wortmannin (bottom panel). Zoom panels in A and B show tail currents that were used for analysis to determine Itail/Itailmax. Recordings at −75 mV are in red. (c) Voltage dependent activation of HCN2 (open symbols, dashed lines) and HCN2 D443N (closed symbols, lines) before (black) and after treatment of the oocytes with Wortmannin (orange, for WT HCN2 n = 7, for HCN2 D443N n = 4). Data points are average ± s.d., lines are fits according to equation 3. Fits for untreated oocytes are from Fig. 4f. Values are summarized in Supplementary Table 1.
Supplementary information
Supplementary Information
Summary of numerical values from TEVC recordings. The data correspond to Fig. 4f,g and Extended Data Fig. 10.
Source data
Source Data Fig. 1
Data for each experimental repeat are given.
Source Data Fig. 2
Data for each experimental repeat are given.
Source Data Fig. 4
Data for each experimental repeat are given.
Source Data Extended Data Fig. 2
Data for each experimental repeat are given.
Source Data Extended Data Fig. 3
Data for each experimental repeat are given.
Source Data Extended Data Fig. 4
Uncropped gel scans are given.
Source Data Extended Data Fig. 10
Data for each experimental repeat are given.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schmidpeter, P.A.M., Wu, D., Rheinberger, J. et al. Anionic lipids unlock the gates of select ion channels in the pacemaker family. Nat Struct Mol Biol 29, 1092–1100 (2022). https://doi.org/10.1038/s41594-022-00851-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00851-2
This article is cited by
-
Constitutive activation mechanism of a class C GPCR
Nature Structural & Molecular Biology (2024)
-
Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel
Nature Communications (2023)
-
Direct regulation of the voltage sensor of HCN channels by membrane lipid compartmentalization
Nature Communications (2023)
-
Assessment of Purity, Functionality, Stability, and Lipid Composition of Cyclofos-nAChR-Detergent Complexes from Torpedo californica Using Lipid Matrix and Macroscopic Electrophysiology
The Journal of Membrane Biology (2023)
-
Gating intermediates reveal inhibitory role of the voltage sensor in a cyclic nucleotide-modulated ion channel
Nature Communications (2022)