Abstract
The regular functioning of the nucleolus and nucleus-mitochondria crosstalk are considered unrelated processes, yet cytochrome c (Cc) migrates to the nucleus and even the nucleolus under stress conditions. Nucleolar liquid–liquid phase separation usually serves the cell as a fast, smart mechanism to control the spatial localization and trafficking of nuclear proteins. Actually, the alternative reading frame (ARF), a tumor suppressor protein sequestered by nucleophosmin (NPM) in the nucleoli, is shifted out from NPM upon DNA damage. DNA damage also triggers early translocation of respiratory Cc to nucleus before cytoplasmic caspase activation. Here, we show that Cc can bind to nucleolar NPM by triggering an extended-to-compact conformational change, driving ARF release. Such a NPM–Cc nucleolar interaction can be extended to a general mechanism for DNA damage in which the lysine-rich regions of Cc—rather than the canonical, arginine-rich stretches of membrane-less organelle components—controls the trafficking and availability of nucleolar proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Dataset generated during the current study is available from the PDB accession code 5EHD. All main data supporting the findings of this study are available within the article, Extended Data and Supplementary Information. Source data are provided with this paper.
References
Di Matteo, A. et al. Molecules that target nucleophosmin for cancer treatment: an update. Oncotarget 7, 44821–44840 (2016).
González-Arzola, K. et al. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Lett. 593, 3101–3119 (2019).
González-Arzola, K. et al. Mitochondrial cytochrome c shot towards histone chaperone condensates in the nucleus. FEBS Open Bio 11, 2418–2440 (2021).
DiGiammarino, E. L., Filippov, I., Weber, J. D., Bothner, B. & Kriwacki, R. W. Solution structure of the p53 regulatory domain of the p19Arf tumor suppressor protein. Biochemistry 40, 2379–2386 (2001).
Sherr, C. J. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer 6, 663–673 (2006).
Yu, Y. et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 26, 3798–3809 (2006).
Grisendi, S., Mecucci, C., Falini, B. & Pandolfi, P. P. Nucleophosmin and cancer. Nat. Rev. Cancer 6, 493–505 (2006).
Box, J. K. et al. Nucleophosmin: from structure and function to disease development. BMC Mol. Biol. 17, 19 (2016).
Korgaonkar, C. et al. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 25, 1258–1271 (2005).
Latonen, L. & Laiho, M. Cellular UV damage responses—functions of tumor suppressor p53. BBA-Rev. Cancer 1755, 71–89 (2005).
Enomoto, T., Lindstrom, M. S., Jin, A., Ke, H. & Zhang, Y. Essential role of the B23/NPM core domain in regulating ARF binding and B23 stability. J. Biol. Chem. 281, 18463–18472 (2006).
Martínez-Fábregas, J. et al. Structural and functional analysis of novel human cytochrome c targets in apoptosis. Mol. Cell. Proteom. 13, 1439–1456 (2014).
Martínez-Fabregas, J. et al. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 5, e1314 (2014).
González-Arzola, K. et al. Structural basis for inhibition of the histone chaperone activity of SET/TAF-Iβ by cytochrome c. Proc. Natl Acad. Sci. USA 112, 9908–9913 (2015).
Rivero-Rodríguez, F. et al. Inhibition of the PP2A activity by the histone chaperone ANP32B is long-range allosterically regulated by respiratory cytochrome c. Redox Biol. 43, 101967 (2021).
Martínez-Fábregas, J. et al. New Arabidopsis thaliana cytochrome c partners: A look into the elusive role of cytochrome c in programmed cell death in plants. Mol. Cell. Proteom. 12, 3666–3676 (2013).
González-Arzola, K. et al. Histone chaperone activity of Arabidopsis thaliana NRP1 is blocked by cytochrome c. Nucleic Acids Res. 45, 2150–2165 (2017).
Goldstein, J. C., Waterhouse, N. J., Juin, P., Evan, G. I. & Green, D. R. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat. Cell Biol. 2, 156–162 (2000).
Nolin, F. et al. Stage-specific changes in the water, Na+, Cl- and K+ contents of organelles during apoptosis, demonstrated by a targeted cryo correlative analytical approach. PLoS ONE 11, e0148727–e0148727 (2016).
Li, P. et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479–489 (1997).
Jacob, S., Miquel, C., Sarasin, A. & Praz, F. Effects of camptothecin on double-strand break repair by non-homologous end-joining in DNA mismatch repair-deficient human colorectal cancer cell lines. Nucleic Acids Res. 33, 106–113 (2005).
Kerppola, T. K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286 (2006).
Qin, F. et al. Knockdown of NPM1 by RNA interference inhibits cells proliferation and induces apoptosis in leukemic cell line. Int. J. Med. Sci. 8, 287–294 (2011).
Mitrea, D. M. et al. Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl Acad. Sci. USA 111, 4466–4471 (2014).
Luchinat, E. et al. Identification of a novel nucleophosmin-interaction motif in the tumor suppressor p14arf. FEBS J. 285, 832–847 (2018).
Lee, H. H. et al. Crystal structure of human nucleophosmin-core reveals plasticity of the pentamer-pentamer interface. Proteins Struct. Funct. Bioinf. 69, 672–678 (2007).
Moreno-Beltrán, B. et al. Cytochrome c1 exhibits two binding sites for cytochrome c in plants. BBA-Bioenerg. 1837, 1717–1729 (2014).
Itahana, K. et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12, 1151–1164 (2003).
Bertwistle, D., Sugimoto, M. & Sherr, C. J. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24, 985–996 (2004).
Freire, E., Schön, A. & Velazquez-Campoy, A. Isothermal titration calorimetry: general formalism using binding polynomials. Methods Enzymol. 455, 127–155 (2009).
Hingorani, K., Szebeni, A. & Olson, M. O. Mapping the functional domains of nucleolar protein B23. J. Biol. Chem. 275, 24451–24457 (2000).
Díaz-Moreno, I., García-Heredia, J. M., Díaz-Quintana, A. & De la Rosa, M. A. Cytochrome c signalosome in mitochondria. Eur. Biophys. J. 40, 1301–1315 (2011).
Moreno-Beltrán, B. et al. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc. Natl Acad. Sci. USA 114, E3041–E3050 (2017).
Elena-Real, C. A. et al. Cytochrome c speeds up caspase cascade activation by blocking 14-3-3ε-dependent Apaf-1 inhibition. Cell Death Dis. 9, 365 (2018).
Elena-Real, C. A. et al. Proposed mechanism for regulation of H2O2-induced programmed cell death in plants by binding of cytochrome c to 14-3-3 proteins. Plant J. 106, 74–85 (2021).
Grummitt, C. G., Townsley, F. M., Johnson, C. M., Warren, A. J. & Bycroft, M. Structural consequences of nucleophosmin mutations in acute myeloid leukemia. J. Biol. Chem. 283, 23326–23332 (2008).
Mitrea, D. M. et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5, e13571 (2016).
Mitrea, D. M. et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 9, 842 (2018).
Gibbs, E., Perrone, B., Hassan, A., Kümmerle, R. & Kriwacki, R. NPM1 exhibits structural and dynamic heterogeneity upon phase separation with the p14ARF tumor suppressor. J. Magn. Reson. 310, 106646 (2020).
Lee, C., Smith, B. A., Bandyopadhyay, K. & Gjerset, R. A. DNA damage disrupts the p14ARF-B23 (nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 65, 9834–9842 (2005).
Nalabothula, N., Indig, F. E. & Carrier, F. The nucleolus takes control of protein trafficking under cellular stress. Mol. Cell Pharmacol. 2, 203–212 (2010).
Wu, M. H., Chang, J. H., Chou, C. C. & Yung, B. Y. Involvement of nucleophosmin/B23 in the response of HeLa cells to UV irradiation. Int. J. Cancer 97, 297–305 (2002).
Yao, Z. et al. B23 acts as a nucleolar stress sensor and promotes cell survival through its dynamic interaction with hnRNPU and hnRNPA1. Oncogene 29, 1821–1834 (2010).
Ukmar-Godec, T. et al. Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat. Commun. 10, 2909 (2019).
Jeng, W.-Y., Chen, C.-Y., Chang, H.-C. & Chuang, W.-J. Expression and characterization of recombinant human cytochrome c in E. coli. J. Bioenerg. Biomembr. 34, 423–431 (2002).
Hu, C.-D., Chinenov, Y. & Kerppola, T. K. Visualization of interactions among bZIP and rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002).
Olteanu, A. et al. Stability and apoptotic activity of recombinant human cytochrome c. Biochem. Biophys. Res. Commun. 312, 733–740 (2003).
Kelley, J. B. & Paschal, B. M. Fluorescence-based quantification of nucleocytoplasmic transport. Methods 157, 106–114 (2019).
Prado Martins, R. et al. In cellulo protein-mRNA interaction assay to determine the action of G-quadruplex-binding molecules. Molecules 23, 3124 (2018).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Greenfield, N. J. Circular dichroism analysis for protein-protein interactions. Methods Mol. Biol. 261, 55–78 (2004).
Hwang, T. L. & Shaka, A. J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A. 112, 275–279 (1995).
Palma, P. N., Krippahl, L., Wampler, J. E. & Moura, J. J. G. BiGGER: a new (soft) docking algorithm for predicting protein interactions. Proteins Struct. Funct. Bioinf. 39, 372–384 (2000).
Pettersen, E. F. et al. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kabsch, W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D. Biol. Crystallogr. 66, 22–25 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. Struct. Biol. 75, 861–877 (2019).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
de la Rosa-Trevín, J. M. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Penczek, P. A. Three-dimensional spectral signal-to-noise ratio for a class of reconstruction algorithms. J. Struct. Biol. 138, 34–46 (2002).
Viranaicken, W. et al. L-Ilf3 and L-NF90 traffic to the nucleolus granular component: alternatively-spliced exon 3 encodes a nucleolar localization motif. PLoS ONE 6, e22296 (2011).
Acknowledgements
We thank the staff at the NMR facility at CITIUS (University of Seville), the microscopy facility at CABIMER (Seville) and the ALBA Synchrotron (Barcelona). We are grateful to the Spanish Government (grant nos. PID2021-126663NB-I00, PGC2018-096049-B-I00/FEDER, BIO2015-70092-R, BFU2015-71017/BMC, BFU2016-75984/BMC, PID2019-105872GB and BFU2017-90030-P, FEDER/Ministerio de Ciencia e Innovación–Agencia Estatal de Investigación), European Regional Development Fund (FEDER), the Regional Government of Andalusia (grant nos. BIO-198; US-1254317, US-1257019, P18-FR-3487 and P18-HO-4091, US/JUNTA/FEDER, UE), the European Commission: European Regional Development Fund and European Research Council (CONCERT, contract number 648201) and the Ramón Areces Foundation. This work has been supported by Infrastructure for NMR, EM and X-rays for Translational Research (iNEXT, grant no. PID 3407) funded by the Horizon 2020 program of the European Commission, Cámara Foundation (C.A.E.R.´s fellowship), the Spanish Ministry of Education, Culture and Sports (grant nos. FPU18/06577, FPU16/01513 and FPU013/04373; M.A.C.-C., A.V.-C. and F.R.-R.’s fellowships, respectively), La Caixa Foundation (E.S.’s fellowship), Severo Ochoa Award of Excellence from MINECO (Government of Spain, IRB Barcelona and CNB Madrid) and the Spanish Association Against Cancer Scientific Foundation (FC AECC, M.P.G.’s postdoctoral grant).
Author information
Authors and Affiliations
Contributions
I.D.-M., M.A.R., K.G.-A., A.D.-Q. and J.M.-F. conceptualized the study. Proteomics and cellular experiments were done by K.G.-A., J.M.-F. and M.A.C.-C. Cloning, expression and purification were done by K.G.-A., C.A.E.-R. and A.V.-C. ITC and SPR measurements were taken by K.G.-A., A.D.-Q., F.R.-R. and A.V.-C. EMSA and circular dichroism spectra were created by K.G.-A. and F.R.-R. NMR data collection and analysis was done by K.G.-A., S.G.-C., I.A. and I.D.-M. Docking calculations were carried out by K.G.-A. Crystallographic data were collected by N.B.-G. and J.A.H. Phase separation assays were done by E.S., X.S., K.G.-A. and A.D.-Q. EM was done by R.A. and J.M.V. PLA analysis was carried out by M.P.G. and R.M.R. Funding was acquired by I.D.-M. and M.A.R. Writing, review and editing of the paper were done by I.D.-M., K.G.-A., M.A.R., A.D.-Q. and J.M.-F.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks Maik Hüttemann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Beth Moorefield and Carolina Perdigoto, in collaboration with the Nature Structural and Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Immunofluorescence analysis of Cc, detection of caspases, γH2AX and p53, along with controls for BIFC assays.
(a) Immunofluorescence analysis of endogenous Cc in Cos-7 cells upon 20 μM CPT or 4 μM Dox treatments for 6 h. Cc was visualized with an anti-Cc antibody (green) using a confocal microscope. Mitochondria were stained with MitoTracker Red CMXRos (red) and nuclei with Hoechst (blue). Non-treated cells were used as control. Co-localization of green Cc fluorescence and blue nuclear staining is shown in the merge images. Scale bars: 10 μm. (b) Left, Detection of caspase-3 in HeLa cells treated with 20 μM CPT for 4, 6, 12, 24 and 48 h. Cleaved caspase-3 (17 kDa) and procaspase-3 (32 kDa) were detected by Western-blot. GADPH (37 kDa) antibody was used as control. Right, Detection of p53 in HeLa cells treated with either 4 μM Dox or 20 μM CPT for 1, 6, 12 and 24 h p53 (53 kDa) and GADPH (37 kDa) were detected. Non-treated cells were used as control. (c) γ-H2AX detection in the nuclei of HeLa cells upon 20 μM CPT treatment for 4 h. Lanes 1 and 2 are for control and CPT-treated samples, respectively. (d) Left, Positive and negative controls for BiFC. HEK293T cells were transfected with the N-terminal YFP fragment (nYFP) attached to bJun (bJun-nYFP), along with the bFos-cYFP vector containing the C-end of YFP bound to bFos (positive control, upper). For negative controls, a construct containing bFos lacking the bJun-interacting domain (bFosΔZip) was used (lower). Right, BiFC analysis of HEK293T cells transfected with casein kinase II subunit β-nYFP (CSNKIIβ-nYFP, upper) or minichromosome maintenance complex 7-nYFP (MCM7-nYFP, lower), along with Cc-cYFP after 6 h of 10 μM CPT treatment. Scale bars: 5 μm. (e) Western-blot analysis vs. anti-Cc (left), anti-NPM (middle) and anti-EGFP (right) antibodies to detect endogenous Cc and NPM, and to confirm protein expression of NPM-nYFP and Cc-cYFP.
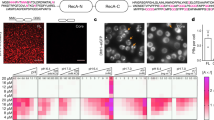
Extended Data Fig. 2 PLA, nucleoli IF controls and quantification of nuclear Cc.
(a) Left, Proximity Ligation Assay (PLA) for Cc:NPM complexes detection in HeLa cells following 20 μM CPT treatment for 6 h. Representative confocal maximal projections of untreated (top) and CPT-treated (bottom) cells, with PLA spots in red and nuclei in blue. As a negative control, we used the rabbit anti-Cc antibody together with an unspecific mouse primary antibody and the two PLA probes (-/+). Scale bar: 25 μm. Right, Scatter plot showing quantification of the PLA signal as the number of PLA spots per nucleus. Data were collected from three independent experiments as that shown in Left. Black lines represent the mean value. The four asterisks (****) stand for p < 0.0001 (p = 1.4 × 10−47 for negative control vs. Cc:NPM; p = 3.8 × 10−46 for Cc:NPM without vs. with CPT), and ‘ns’ (p = 0.05793) for statistically not significant (one-way ANOVA followed by a Tukey´s post hoc test). (b) Immunofluorescence analysis of endogenous Cc in HeLa cells, upon 20 μM CPT treatment for 6 h. Cc was visualized with an anti-Cc antibody (green), whereas Pol I was immunodetected with an anti-Pol I antibody (red). Nuclei were stained with Hoechst (blue). Non-treated cells were used as control. Co-localization of green Cc fluorescence and red Pol I fluorescence is shown in the merged images. Scale bars: 5 μm. (c) Quantification of nuclear Cc (%), obtained as the percentage of the Cc total fluorescence intensity in the nucleus of HeLa cells respecting to that for the whole cell. Fluorescence intensity was quantified in 48 nontreated (control), 43 CPT-treated and 45 Dox-treated cells. Black lines represent the mean value. The four asterisks (****) stand for p < 0.0001 (p = 5.0 × 10−19 for control vs. CPT, p = 4.3 × 10−11 for control vs. Dox), and ‘ns’ (p = 0.14659) for statistically not significant. One-way ANOVA statistics with Tukey´s test were performed.
Extended Data Fig. 3 Effect of the His-tag on NPM(1-130) assesed by NMR, CD and SDS-PAGE.
(a, b) Overlay of the structures of NPM core with26 (PDB code 2P1B; beige) and without24 (PDB code 4N8M; cyan) a 6x-His tag at the C-end. (a) Richardson’s ribbon diagrams. (b) Full backbone and main-chain atoms in the upper and lower views, respectively. (c) Snapshot of the sequence alignment of the two proteins as shown in UCSF Chimera. Green shadows highlight β strands. Pale yellow background corresponds to helical (α) regions. Red empty boxes remark regions for which structural data is missing in the PDB files. Residue symbols are colored according to the Clustal-X pattern. (d) Two-dimensional transverse relaxation-optimized spectroscopy (2D TROSY) spectrum of His-tagged 2H-15N-labeled NPM(1-130) (50 µM, pentamer) in 10 mM potassium phosphate pH 7.0 containing 1 mM TCEP, as recorded at a Bruker Avance 700 MHz NMR spectrometer at 298 K. (e) CD spectra of 3 µM His-tagged NPM(1-130) (pentamer) in 10 mM potassium phosphate pH 7.0 with or without 150 mM NaCl. (f) SDS-PAGE of non-labeled NPM(1-130) with (left) or without (right) a His-tag, under low salt (-NaCl) or high salt (+NaCl) concentrations. Those bands matching the molecular mass of the pentamer or monomer are indicated as *P or *M, respectively. Molecular weight markers are on the left lane at each panel.
Extended Data Fig. 4 Folding of NPM constructs, competition assay between Cc and ARF for NPM(9-122) and EMSA between NPM(1-130) and Cc.
(a) Far-UV CD spectra of wild-type (WT) NPM constructs NPM(1-294), NPM(9-122), NPM(225-294) and NPM(123-294), and NPM(1-294) mutants Q15A, E37K and E37K/N38A. (b) SDS-PAGE of WT NPM(1-294) (lane 1) and Q15A, E37K and E37K/N38A mutants (lanes 2-4, respectively). The band matching the molecular mass of the pentamer is indicated as *P. Molecular weight markers are on the left lane. (c) Competition between Cc and p19ARF for NPM(9-122) binding. 1D 1H NMR spectra monitoring the Met80-methyl signal of 13 µM reduced Cc, either free (solid black) or upon successive additions of 26 µM NPM(9-122) (dotted line) and the p19ARF peptide at increasing concentrations of 40 µM (yellow), 50 µM (red), 52 µM (magenta), 55 µM (blue) and 57 µM (brown). (d) EMSA showing the interaction between NPM(1-130) and Cc. Migration of NPM(1-130) either free or in complex with Cc are indicated. With the only exception of NPM(225-294) and NPM(123-294) constructs, NPM molar concentration corresponds to the pentameric form.
Extended Data Fig. 5 Thermodynamic analysis of the Cc interaction with NPM constructs and mutants.
ITC titrations of reduced Cc with wild-type (WT) NPM(1-294) (a), NPM(9-122) (b), NPM(225-294) (c), NPM(123-294) (d), 6xHis-tagged NPM(1-130) (e), NPM(1-130) without 6xHis-tag (f), Q15A NPM(1-294) (g), E37K NPM(1-294) (h) and E37K/N38A NPM(1-294) (i). Thermograms (upper plots) and binding isotherms (lower plots) were obtained at low ionic conditions, except for ITC titrations shown in (e) and (f), in which 0.1 M KCl was added. With the only exception of NPM(225-294) and NPM(123-294) constructs, NPM molar concentration corresponds to the pentameric form.
Extended Data Fig. 6 Effect of salt and redox state on ITC titrations, SPR and CD measurements of Cc and NPM interactions.
(a, b) ITC titrations of Cc with WT NPM(1-294) and WT NPM(9-122) in the presence of 0.1 M KCl. (c, d) ITC titrations of oxidized Cc with WT NPM(1-294) and WT NPM(9-122) at low ionic conditions. Thermograms (upper) and binding isotherms (lower) are shown. (e) SPR sensorgrams for the binding of Cc to NPM(1-294) (upper plot) or NPM(9-122) (lower plot). NPM(1-294) concentrations were 0.1, 0.5 and 1.0 μM, whereas NPM(9-122) concentrations were 1, 5 and 10 μM. Three replicate injections were performed for each protein concentration. In each sensorgram, the signals from the control surface were subtracted. Kinetic dissociation constant (koff) values are shown. (f) Changes in the far-UV (left plots) and visible (right plots) CD spectra of Cc upon WT NPM(9-122) binding. Spectra of the isolated proteins—Cc (red lines) and NPM(9-122) (black lines)—along with the Cc:NPM(9-122) complex at the 1:1 ratio (green lines) and the sum of the spectra of the isolated proteins (blue lines) are shown. Insets: difference between the spectra of the complex (green lines) and the sum of the isolated proteins (blue lines). NPM molar concentration corresponds to the pentameric form.
Extended Data Fig. 7 ITC titrations of p19ARF with NPM, CSPs and line-width analyses of Cc-NPM(9-122) complex.
(a, b) ITC titration of the p19ARF(1-37) peptide with NPM(1-294) (a) and NPM(9-122) (b) at low ionic strength. (c) ITC titration of the p19ARF(83-102) peptide with NPM(1-294) at low ionic strength. Thermograms (upper plots) and binding isotherms (lower plots) are shown. (d, e) NMR titrations of 15N-labeled Cc with NPM(9-122). (d) Average CSPs (ΔδAvg) experienced by amide resonances of Cc in complex with NPM(9-122) at the Cc:NPM ratio of 1:0.5. (e) Differences in the line-width (ΔΔν1/2Binding) of 15N dimension of the NMR amide signals of Cc upon binding to NPM(9-122). The ΔΔν1/2Binding values were calculated from the difference between [1H-15N] HSQC spectra of free Cc and those of the complex at the above molar ratio. The threshold (dashed line) corresponds to the average plus two times the standard deviation (ΔΔν1/2Binding ≥ ΔΔν1/2Binding + 2Sn-1). Residues beyond the threshold exhibit substantial line broadening. NPM molar concentration corresponds to the pentameric form.
Extended Data Fig. 8 NMR analysis of 2H-15N-labeled NPM(1-130) with either Cc or ARF.
(a, b) NMR titrations of 2H-15N-labeled NPM(1-130) with either Cc at the Cc:NPM(1-130) molar ratio of 2:1 (a) or p19ARF(1-37) at the p19ARF(1-37):NPM(1-130) molar ratio of 3:1 (b). Free and bound NPM(1-130) spectra are represented in blue and red, respectively. Backbone assignments of well-dispersed NPM(1-130) resonances are shown. (c, d) Average CSPs (ΔδAvg) experienced by amide NMR signals of NPM(1-130) after binding to either Cc (c) or p19ARF(1-37) (d) as determined at the above molar ratios. The dashed lines correspond to the average plus one or two times the standard deviation. (e, f) Intensity variation experienced by amide NMR signals of NPM(1-130) upon binding to either Cc (e) or p19ARF(1-37) (f), expressed as the ratio of resonance intensities of complexed NPM(1-130) over the corresponding resonance intensity of free NPM(1-130). The dashed lines stand for the average calculated after excluding the five largest and five smallest intensity values. NPM molar concentration corresponds to the pentameric form.
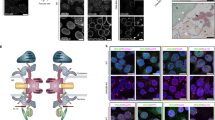
Extended Data Fig. 9 Electron-density map of NPM(9-122), electron micrographs and molecular docking of Cc:NPM(9-122).
(a) Left. Electron-density map (2Fo-Fc map contoured at 1σ) for one of the NPM(9-122) pentamers determined in the presence of Cc (this work). Right. Immunoblotting against NPM and Cc in the non-diluted complex Cc:NPM(9-122), the diluted Cc:NPM(9-122) complexes (ranging from 1:10 to 1:1000) and in the extensively washed crystals of the same complex. One asterisk stands for the NPM pentamer (67.6 kDa), whereas two asterisks indicate an NPM decamer (135.2 kDa). (b-d) Negatively stained electron micrographs of NPM(1-294) (b), Cc:NPM(1-294) (c) and Cc:NPM(9-122) (d), from which three-dimensional reconstructions were carried out. In (b) front and side views are indicated by arrows and arrowheads, respectively. Scale bar is 50 nm. (e) NMR-based BiGGER molecular docking of the Cc:NPM(9-122) complex. Ribbon representation corresponds to the best complex model inferred from global calculation scores. NPM(9-122) and Cc ribbons are shown in beige and red respectively, the Cc heme group is in green. The structural PDB coordinates of NPM(9-122) (PDB code 5EHD, this work) and Cc (PDB code 1J3S45) were used as inputs for docking calculations. NPM molar concentration corresponds to the pentameric form.
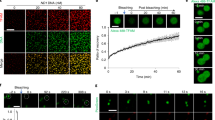
Extended Data Fig. 10 LLPS of NPM(1-294) with either ARF or Cc, effect of Cc addition on isolated nucleoli over time, and FRAP curves of ARF-EGFP.
(a) In vitro droplets formed by NPM(1-294) with either p19ARF(1-37) (left) or Cc (right) showed by DIC microscopy images. NPM(1-294) (pentamer) was at 5 μM, whereas p19ARF(1-37) and Cc were added at p19ARF(1-37)-NPM(1-294) and Cc-NPM(1-294) ratios of 5:1 and 10:1, respectively. (b) In vitro droplets formed by NPM(1-294) (pentamer) with either p19ARF or Cc in 10 mM sodium phosphate buffer (pH 7.4) containing 100 mM KCl. The DIC microscopy images show phase separation induced by NPM(1-294) with p19ARF(1-37) (upper) or Cc (lower). NPM(1-294) was used at 5 μM, whereas p19ARF(1-37) and Cc were added at p19ARF(1-37):NPM(1-294) and Cc:NPM(1-294) ratios of 5:1 and 10:1. (c) Confocal microscopy images of isolated nucleoli pre-incubated with 5 μM NPM(1-294) and 5 μM p19ARF(1-37) upon addition of 5 μM Cc over time. Scale bars are 3 μM. All images were taken with a 63x objective (oil). (d) Kinetics of p19ARF-EGFP fluorescence recording emitted from the bleached region (yellow square) in Fig. 7c is represented by a red solid line; a non-bleached region of a different nucleolus is denoted by a red dotted line; a nucleoplasm delimited region is represented by a black dotted line, and a region outside the nucleus is depicted by a black dashed line. Cells over-expressed p19ARF-EGFP and NPM-mCherry (Fig. 7c). (e) Kinetics of p19ARF-EGFP fluorescence recovery (see Fig. 7d, yellow boxes) emitted from the bleached region (blue solid line), a non-bleached region of a different nucleolus (blue dotted line), a nucleoplasm delimited region (black dotted line) and a region outside the nucleus (black dashed line). Cells over-expressed p19ARF-EGFP, NPM-mCherry and Cc-AmCyan (Fig. 7d). (f) FRAP curves of p19ARF-EGFP fluorescence recovery kinetics of cells expressing p19ARF-EGFP and NPM-mCherry (red line) or p19ARF-EGFP, NPM-mCherry and Cc-AmCyan (blue line), after normalization of fluorescence intensities. Error bars are standard deviations of the mean (SEM) of 12 to 15 cells.
Supplementary information
Supplementary Information
Supplementary Notes, References and Table 1.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed gel.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed gels.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
González-Arzola, K., Díaz-Quintana, A., Bernardo-García, N. et al. Nucleus-translocated mitochondrial cytochrome c liberates nucleophosmin-sequestered ARF tumor suppressor by changing nucleolar liquid–liquid phase separation. Nat Struct Mol Biol 29, 1024–1036 (2022). https://doi.org/10.1038/s41594-022-00842-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00842-3
This article is cited by
-
Diverse functions of cytochrome c in cell death and disease
Cell Death & Differentiation (2024)
-
Apoptotic proteins with non-apoptotic activity: expression and function in cancer
Apoptosis (2023)