Abstract
In addition to its role in chromosome maintenance, the six-membered Smc5/6 complex functions as a restriction factor that binds to and transcriptionally silences viral and other episomal DNA. However, the underlying mechanism is unknown. Here, we show that transcriptional silencing by the human Smc5/6 complex is a three-step process. The first step is entrapment of the episomal DNA by a mechanism dependent on Smc5/6 ATPase activity and a function of its Nse4a subunit for which the Nse4b paralog cannot substitute. The second step results in Smc5/6 recruitment to promyelocytic leukemia nuclear bodies by SLF2 (the human ortholog of Nse6). The third step promotes silencing through a mechanism requiring Nse2 but not its SUMO ligase activity. By contrast, the related cohesin and condensin complexes fail to bind to or silence episomal DNA, indicating a property unique to Smc5/6.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data are provided with this paper. All other data are available from corresponding authors on reasonable request.
Change history
10 May 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41594-023-01009-4
References
Jeppsson, K., Kanno, T., Shirahige, K. & Sjögren, C. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 15, 601–614 (2014).
Gligoris, T. & Löwe, J. Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol. 26, 680–693 (2016).
Alt, A. et al. Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat. Commun. 8, 14011 (2017).
Adamus, M. et al. Molecular Insights into the architecture of the human SMC5/6 complex. J. Mol. Biol. 432, 3820–3837 (2020).
Palecek, J., Vidot, S., Feng, M., Doherty, A. J. & Lehmann, A. R. The Smc5-Smc6 DNA repair complex: bridging of the Smc5-Smc6 heads by the kleisin, nse4, and non-kleisin subunits. J. Biol. Chem. 281, 36952–36959 (2006).
Kanno, T., Berta, D. G. & Sjögren, C. The Smc5/6 complex is an ATP-dependent intermolecular DNA linker. Cell Rep. 12, 1471–1482 (2015).
Aragón, L. The Smc5/6 complex: new and old functions of the enigmatic long-distance relative. Annu. Rev. Genet. 52, 89–107 (2018).
Palecek, J. J. SMC5/6: multifunctional player in replication. Genes 10, 7 (2019).
Palecek, J. J. & Gruber, S. Kite proteins: a superfamily of SMC/kleisin partners conserved across bacteria, archaea, and eukaryotes. Structure 23, 2183–2190 (2015).
De Piccoli, G. et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 8, 1032–1034 (2006).
Potts, P. R., Porteus, M. H. & Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25, 3377–3388 (2006).
Ampatzidou, E., Irmisch, A., O’Connell, M. J. & Murray, J. M. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 26, 9387–9401 (2006).
Betts Lindroos, H. et al. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22, 755–767 (2006).
Menolfi, D., Delamarre, A., Lengronne, A., Pasero, P. & Branzei, D. Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol. Cell 60, 835–846 (2015).
Li, X. & Heyer, W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113 (2008).
Bermúdez-López, M. et al. The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 38, 6502–6512 (2010).
Kegel, A. & Sjögren, C. The Smc5/6 complex: more than repair? Cold Spring Harb. Symp. Quant. Biol. 75, 179–187 (2010).
Jeppsson, K. et al. The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 10, e1004680 (2014).
Kegel, A. et al. Chromosome length influences replication-induced topological stress. Nature 471, 392–396 (2011).
Decorsière, A. et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 531, 386–389 (2016).
Murphy, C. M. et al. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep. 16, 2846–2854 (2016).
Li, T., Robert, E. I., van Breugel, P. C., Strubin, M. & Zheng, N. A promiscuous α-helical motif anchors viral hijackers and substrate receptors to the CUL4–DDB1 ubiquitin ligase machinery. Nat. Struct. Mol. Biol. 17, 105–112 (2010).
Abdul, F. et al. Smc5/6 antagonism by HBx is an evolutionarily conserved function of hepatitis B virus infection in mammals. J. Virol. 92, e00769-18 (2018).
van Breugel, P. C. et al. Hepatitis B virus X protein stimulates gene expression selectively from extrachromosomal DNA templates. Hepatology 56, 2116–2124 (2012).
Dupont, L. et al. The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagonized by Vpr. Cell Host Microbe 29, 792–805.e6 (2021).
Gibson, R. T. & Androphy, E. J. The SMC5/6 complex represses the replicative program of high-risk human papillomavirus type 31. Pathogens 9, 786 (2020).
Nagy, G. et al. Motif oriented high-resolution analysis of ChIP-seq data reveals the topological order of CTCF and cohesin proteins on DNA. BMC Genomics 17, 637 (2016).
Sutani, T. et al. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat. Commun. 6, 7815 (2015).
Taylor, E. M., Copsey, A. C., Hudson, J. J. R., Vidot, S. & Lehmann, A. R. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol. Cell. Biol. 28, 1197–1206 (2008).
Harvey, S. H., Krien, M. J. & O’Connell, M. J. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol. 3, reviews3003.1 (2002).
Arumugam, P. et al. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr. Biol. 13, 1941–1953 (2003).
Hirano, M., Anderson, D. E., Erickson, H. P. & Hirano, T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 20, 3238–3250 (2001).
Guerineau, M. et al. Analysis of the Nse3/MAGE-binding domain of the Nse4/EID family proteins. PLoS ONE 7, e35813 (2012).
Hudson, J. J. R. et al. Interactions between the Nse3 and Nse4 components of the SMC5-6 complex identify evolutionarily conserved interactions between MAGE and EID families. PLoS ONE 6, e17270 (2011).
Vondrova, L. et al. A role of the Nse4 kleisin and Nse1/Nse3 KITE subunits in the ATPase cycle of SMC5/6. Sci. Rep. 10, 9694 (2020).
Venegas, A. B., Natsume, T., Kanemaki, M. & Hickson, I. D. Inducible degradation of the human SMC5/6 complex reveals an essential role only during interphase. Cell Rep. 31, 107533 (2020).
Jo, A., Li, S., Shin, J. W., Zhao, X. & Cho, Y. Structure basis for shaping the Nse4 protein by the Nse1 and Nse3 dimer within the Smc5/6 complex. J. Mol. Biol. 433, 166910 (2021).
Zabrady, K. et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res. 44, 1064–1079 (2016).
Duan, X. et al. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol. Cell 35, 657–668 (2009).
Solé-Soler, R. & Torres-Rosell, J. Smc5/6, an atypical SMC complex with two RING-type subunits. Biochem. Soc. Trans. 48, 2159–2171 (2020).
Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA 102, 4777–4782 (2005).
Jacome, A. et al. NSMCE2 suppresses cancer and aging in mice independently of its SUMO ligase activity. EMBO J. 34, 2604–2619 (2015).
Andrews, E. A. et al. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25, 185–196 (2005).
Pond, K. W., de Renty, C., Yagle, M. K. & Ellis, N. A. Rescue of collapsed replication forks is dependent on NSMCE2 to prevent mitotic DNA damage. PLoS Genet. 15, e1007942 (2019).
Potts, P. R. & Yu, H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25, 7021–7032 (2005).
Zapatka, M. et al. Sumoylation of Smc5 promotes error-free bypass at damaged replication forks. Cell Rep. 29, 3160–3172.e4 (2019).
Boulanger, M., Chakraborty, M., Tempé, D., Piechaczyk, M. & Bossis, G. SUMO and transcriptional regulation: the lessons of large-scale proteomic, modifomic and genomic studies. Molecules 26, 828 (2021).
Ni, H. J. et al. Depletion of SUMO ligase hMMS21 impairs G1 to S transition in MCF-7 breast cancer cells. Biochim. Biophys. Acta 1820, 1893–1900 (2012).
Niu, C. et al. The Smc5/6 complex restricts HBV when localized to ND10 without inducing an innate immune response and is counteracted by the HBV X protein shortly after infection. PLoS ONE 12, e0169648 (2017).
Potts, P. R. & Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14, 581–590 (2007).
Pebernard, S., Wohlschlegel, J., McDonald, W. H., Yates, J. R. 3rd & Boddy, M. N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 26, 1617–1630 (2006).
Yu, Y. et al. Integrative analysis reveals unique structural and functional features of the Smc5/6 complex. Proc. Natl Acad. Sci. USA 118, e2026844118 (2021).
Räschle, M. et al. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671 (2015).
Bustard, D. E. et al. During replication stress, non-Smc element 5 (Nse5) is required for Smc5/6 protein complex functionality at stalled forks. J. Biol. Chem. 287, 11374–11383 (2012).
Leung, G. P., Lee, L., Schmidt, T. I., Shirahige, K. & Kobor, M. S. Rtt107 is required for recruitment of the SMC5/6 complex to DNA double strand breaks. J. Biol. Chem. 286, 26250–26257 (2011).
Oravcová, M. et al. Brc1 promotes the focal accumulation and SUMO ligase activity of Smc5-Smc6 during replication stress.Mol. Cell. Biol. 39, e00271-18 (2019).
Etheridge, T. J. et al. Live-cell single-molecule tracking highlights requirements for stable Smc5/6 chromatin association in vivo. eLife 10, e68579 (2021).
Gutierrez-Escribano, P. et al. Purified Smc5/6 complex exhibits DNA substrate recognition and compaction. Mol. Cell 80, 1039–1054.e6 (2020).
Wilhelm, L. et al. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4, e06659 (2015).
Hu, B. et al. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr. Biol. 21, 12–24 (2011).
Serrano, D. et al. The Smc5/6 core complex is a structure-specific DNA binding and compacting machine. Mol. Cell 80, 1025–1038.e5 (2020).
Hu, B. et al. Qri2/Nse4, a component of the essential Smc5/6 DNA repair complex. Mol. Microbiol. 55, 1735–1750 (2005).
Båvner, A., Matthews, J., Sanyal, S., Gustafsson, J.-Å. & Treuter, E. EID3 is a novel EID family member and an inhibitor of CBP-dependent co-activation. Nucleic Acids Res. 33, 3561–3569 (2005).
Corpet, A. et al. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res. 48, 11890–11912 (2020).
Bauer, B. W. et al. Cohesin mediates DNA loop extrusion by a ‘swing and clamp’ mechanism. Cell 184, 5448–5464.e22 (2021).
Bürmann, F. et al. A folded conformation of MukBEF and cohesin. Nat. Struct. Mol. Biol. 26, 227–236 (2019).
Lee, B. G. et al. Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat. Struct. Mol. Biol. 27, 743–751 (2020).
Varejão, N. et al. Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs. Nat. Commun. 12, 7013 (2021).
Bermúdez-López, M. et al. ATPase-dependent control of the Mms21 SUMO ligase during DNA repair. PLoS Biol. 13, e1002089 (2015).
Everett, R. D. The spatial organization of DNA virus genomes in the nucleus. PLoS Pathog. 9, e1003386 (2013).
Michelini, Z., Negri, D. & Cara, A. Integrase defective, nonintegrating lentiviral vectors. Methods Mol. Biol. Clifton NJ 614, 101–110 (2010).
Cuchet-Lourenço, D., Vanni, E., Glass, M., Orr, A. & Everett, R. D. Herpes simplex virus 1 ubiquitin ligase ICP0 interacts with PML isoform I and induces its SUMO-independent degradation. J. Virol. 86, 11209–11222 (2012).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Acknowledgements
We thank T. Gligoris and S. Gruber for help in designing the Smc5 and Smc6 ATPase mutants; O. Fernandez-Capetillo for providing the anti-Nse2 antibody; and S. Kassem and J. Curran for critical reading of the manuscript. This work was supported by grants from the Swiss National Science Foundation (310030-149626 and 310030-175781) to M.S., by the Canton of Geneva and by Gilead Sciences.
Author information
Authors and Affiliations
Contributions
F.A., R.K.B., S.P.F. and M.S. conceived the study and designed the experiments. D.R. performed the PHH experiments shown in Extended Data Fig. 1a,b. D.R. and D.K. performed the confocal microscopy experiments shown in Fig. 4d. R.K.B. and D.K. performed the confocal microscopy experiments shown in Fig. 5d. F.A. performed all the other experiments. B.B. contributed to several figures. A.D. contributed to the revision of the manuscript. All authors interpreted the results. F.A. and M.S. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
This study was partly funded by Gilead Sciences, Inc., and D.R., D.K., R.K.B. and S.P.F are employees of Gilead Sciences, Inc. The other authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Camilla Björkegren and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Beth Moorefield and Carolina Perdigoto, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
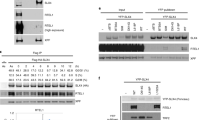
Extended Data Fig. 1 Smc5/6 is the only SMC complex to silence expression of the episomal HBV genome. Related to Fig. 1.
a, PHH were transfected with a non-targeting control siRNA (siCtrl) or with siRNA targeting the indicated Smc subunits. Cells were then incubated for 3 days and infected with wild-type (WT) or an HBx-deficient (ΔX) HBV. HBeAg secretion, a marker for HBV gene expression, was measured 14 days later. HBeAg levels are expressed as a percentage of those in control cells infected with HBV(WT) (grey bars) or in siSmc6-treated cells infected with HBV(ΔX) (black bars). Data are means ± SEM of three independent experiments with samples from one PHH donor. The schematic diagram summarizing the design of the study is shown at the top. b, The mRNA levels of the indicated Smc proteins in the samples analyzed in a were determined by real-time RT–PCR and normalized to β-actin. Values are expressed as a percentage of those in control cells (siCtrl). Data are means ± SEM of three independent experiments with one PHH donor. c, Luciferase assay for the ChIP experiment presented in Fig, 1c. Data are means ± SEM of 2 independent experiments. d, HA-Smc2 assembles into a condensin complex. Whole extracts prepared from HepG2 cells expressing the indicated HA-tagged SMC protein in a CRISPR/Cas9 knockout background were immunoprecipitated with anti-HA antibodies. The amounts recovered and the presence of Smc5, CAP-H and Nse4 in the eluates were assessed by Western blotting. CAP-H and Nse4 are the kleisin subunits of, respectively, condensin (Smc2) and the Smc5/6 complex. See Fig. 1a. Actin serves as a negative control. The experiment was repeated twice independently with similar results.
Extended Data Fig. 2 Effect of single-subunit depletion on Smc5/6 complex integrity and restriction activity.
HepG2 cells were transduced with lentiviral constructs expressing Cas9 alone (Ctrl) or Cas9 together with sgRNA targeting the indicated Smc5/6 subunits, or with lentiviruses encoding GFP or GFP-HBx, as indicated. After selection, cells were transfected with a luciferase reporter plasmid. Luciferase assay and Western blot analysis were performed 5 days later. Hsp90 serves as a loading control. Unessential lanes were removed from the original blot images. Luciferase activity is relative to that measured in cells expressing HBx, which was set to 10. Data are means ± SEM of 3 independent experiments.
Extended Data Fig. 3 Involvement of Smc5 and Smc6 ATP binding and hydrolysis in Smc5/6 episomal DNA binding and restriction, and degradation by HBx. Related to Fig. 2.
a, Luciferase assay for the ChIP experiment presented in Fig. 2c. Data are means ± SEM of 3 independent experiments. b, Smc6 ATP binding mutants normally assemble into Smc5/6 complexes. Whole extracts prepared from HepG2 cells depleted of Smc6 and expressing HA-GFP or the indicated HA-tagged SMC protein were immunoprecipitated with anti-HA antibodies. The amounts recovered and the presence of other Smc5/6 subunits in the eluates were assessed by Western blotting. Hsp90 serves as a negative control. The experiment was repeated twice independently with similar results. c, Luciferase assay for the ChIP experiment presented in Fig, 2d. Data are means ± SEM of 3 independent experiments. d, HepG2 cells depleted of Smc6 and expressing GFP or the indicated wild-type or mutant Smc6 proteins from lentiviral vectors exactly as in Fig. 2b were also transduced with GFP or GFP-HBx. Western blot analysis was performed as before. GAPDH serves as a loading control. The experiment was repeated thrice independently with similar results.
Extended Data Fig. 4 Nse4a performs an essential function in Smc5/6 restriction for which Nse4b cannot substitute. Related to Fig. 3.
a, Schematic diagram of Nse4a (long), the shorter isoform of Nse4a, Nse4b and the Nse4b variant bearing the N-terminal region unique to Nse4a. White boxes indicate Nse4a sequences and the region of the short Nse4a protein common to both splicing isoforms. Hatched boxes indicate regions of Nse4b showing homology to Nse4a. The grey box indicates a region with no homology. Highlighted in black are the highly conserved N-terminal and C-terminal kleisin domains that have the potential to form helix-turn-helix and winged-helix motifs and are involved in Nse4 interaction with Smc6 and Smc5, respectively (Palecek et al., 2006; Vondrova et al., 2020). b, Luciferase assay for the ChIP experiment presented in Fig. 3b. Data are means ± SEM of 2 independent experiments. c, Same experiment as in Fig. 3a but including an Nse4b chimeric protein carrying the N-terminal region unique to Nse4a (Nse4a-b; see panel a). Expression of Nse4b and Nse4a-b was inferred from their stabilization effect on the other Smc5/6 subunits (lane 8). Data are means ± SEM of 2 independent experiments.
Extended Data Fig. 5 Nse1 and Nse3 DNA-binding mutants are functional in vivo.
a,b, Control HepG2 cells (black bars) and cells depleted of Nse1 (a) or Nse3 (b; grey bars) were transfected with a luciferase reporter plasmid and shortly after transduced with GFP or the corresponding wild-type or DNA-binding mutant protein as indicated (Zabrady et al., 2016). Luciferase assay and Western blot analysis were as before. Data are means ± SEM of 3 independent experiments.
Extended Data Fig. 6 HBx triggers Smc5/6 degradation in the absence of Nse2. Related to Fig. 4.
a, Control HepG2 cells (black bars) and cells depleted of Nse2 (grey bars) were transfected with a reporter gene and transduced with GFP or Flag-tagged Nse2 (F-Nse2). Cells were then split and further transduced with GFP or GFP-HBx. Luciferase assay and Western blot analysis were as before. Data are expressed as mean ± SEM of 2 independent experiments. b, Luciferase assay for the ChIP experiment presented in Fig. 4b. Data are expressed as mean ± SEM of 3 independent experiments. c, Single channel confocal images of middle right panels merged images presented in Fig. 4d.
Extended Data Fig. 7 HBx triggers Smc5/6 degradation in the absence of SLF2. Related to Fig. 5.
a, Control HepG2 cells (black bars) and cells depleted of SLF2 (grey bars) were transfected with a reporter gene and shortly after transduced with GFP, GFP-HBx and/or SLF2 in the indicated combinations. Luciferase assay and Western blot analysis were performed as before. Data are expressed as mean ± SEM of 2 independent experiments. b, Luciferase assay for the ChIP experiment presented in Fig. 5b. Data are means ± SEM of 2 independent experiments. c, Single channel confocal images of middle right panels merged images presented in Fig. 5d. d, Luciferase assay for the ChIP experiment presented in Fig. 5e. Data are means ± SEM of 4 independent experiments.
Supplementary information
Source data
Source Data Fig. 1
Source data of graphical representations used in Fig. 1.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Source data of graphical representations used in Fig. 2.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Source data of graphical representations used in Fig. 3.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Source data of graphical representations used in Fig. 4.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Source data of graphical representations used in Fig. 5.
Source Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 1
Source data of graphical representations used in Extended Fig. 1.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Source data of graphical representations used in Extended Fig. 2.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Source data of graphical representations used in Extended Fig. 3.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Source data of graphical representations used in Extended Fig. 4.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Source data of graphical representations used in Extended Fig. 5.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Source data of graphical representations used in Extended Fig. 6.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Source data of graphical representations used in Extended Fig. 7.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abdul, F., Diman, A., Baechler, B. et al. Smc5/6 silences episomal transcription by a three-step function. Nat Struct Mol Biol 29, 922–931 (2022). https://doi.org/10.1038/s41594-022-00829-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00829-0
This article is cited by
-
The cell biology of HIV-1 latency and rebound
Retrovirology (2024)
-
The interactions between PML nuclear bodies and small and medium size DNA viruses
Virology Journal (2023)
-
Genome control by SMC complexes
Nature Reviews Molecular Cell Biology (2023)