Abstract
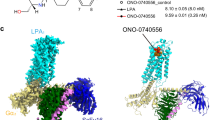
Lysophosphatidylcholine (LPC) is an essential mediator in human lipid metabolism and is associated with a variety of diseases, but the exact identity of LPC receptors remains controversial. Through extensive biochemical and structural analyses, we have identified the orphan receptor GPR119 as the receptor for LPC. The structure of the GPR119–G-protein complex without any added ligands reveals a density map that fits well with LPC, which is further confirmed by mass spectrometry and functional studies. As LPCs are abundant on the cell membrane, their preoccupancy in the receptor may lead to ‘constitutive activity’ of GPR119. The structure of GPR119 bound to APD668, a clinical drug candidate for type 2 diabetes, reveals an exceedingly similar binding mode to LPC. Together, these data highlight structural evidence for LPC function in regulating glucose-dependent insulin secretion through direct binding and activation of GPR119, and provide structural templates for drug design targeting GPR119.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Materials are available from the corresponding authors upon reasonable request. Density maps and structure coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the PDB with accession codes EMD-33525 and PDB 7XZ5 for the LPC-GPR119–Gs complex; EMD-33526 and PDB 7XZ6 for the APD668–GPR119–Gs complex. Source data are provided with this paper. All other relevant data are available from the corresponding author upon request.
References
Tsukahara, T., Matsuda, Y. & Haniu, H. Lysophospholipid-related diseases and PPARgamma signaling pathway. Int. J. Mol. Sci. https://doi.org/10.3390/ijms18122730 (2017).
Wilson, P. W., D’Agostino, R. B., Parise, H., Sullivan, L. & Meigs, J. B. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation 112, 3066–3072 (2005).
Yki-Järvinen, H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. 2, 901–910 (2014).
Liu, P. et al. The mechanisms of lysophosphatidylcholine in the development of diseases. Life Sci. 247, 117443 (2020).
Zhao, Z. et al. Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. J. Clin. Oncol. 25, 2696–2701 (2007).
Munder, P. G., Modolell, M., Andreesen, R., Weltzien, H. U. & Westphal, O. Lysophosphatidylcholine (lysolecithin) and its synthetic analogues. Immunemodulating and other biologic effects. Springe. Semin. Immunopathol. 2, 187–203 (1979).
Knuplez, E. & Marsche, G. An updated review of pro- and anti-inflammatory properties of plasma lysophosphatidylcholines in the vascular system. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21124501 (2020).
Matsumoto, T., Kobayashi, T. & Kamata, K. Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr. Med. Chem. 14, 3209–3220 (2007).
Soga, T. et al. Lysophosphatidylcholine enhances glucose-dependent insulin secretion via an orphan G-protein-coupled receptor. Biochem. Biophys. Res. Commun. 326, 744–751 (2005).
Drzazga, A. et al. Lysophosphatidylcholine and its phosphorothioate analogues potentiate insulin secretion via GPR40 (FFAR1), GPR55 and GPR119 receptors in a different manner. Mol. Cell. Endocrinol. 472, 117–125 (2018).
Chu, Z.-L. et al. A role for β-cell-expressed G protein-coupled receptor 119 in glycemic control by enhancing glucose-dependent insulin release. Endocrinology 148, 2601–2609 (2007).
Chu, Z.-L. et al. A role for intestinal endocrine cell-expressed g protein-coupled receptor 119 in glycemic control by enhancing glucagon-like Peptide-1 and glucose-dependent insulinotropic Peptide release. Endocrinology 149, 2038–2047 (2008).
Hothersall, J. D. et al. Sustained wash-resistant receptor activation responses of GPR119 agonists. Eur. J. Pharmacol. 762, 430–442 (2015).
Alexander, S. P. et al. Hydrolases (version 2019.5) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHAR/BPS Guide to Pharmacology CITE https://doi.org/10.2218/gtopdb/F799/2019.5 (2019).
Overton, H., Fyfe, M. & Reynet, C. GPR119, a novel G protein‐coupled receptor target for the treatment of type 2 diabetes and obesity. Brit J. Pharm. 153, S76–S81 (2008).
Lauffer, L. M., Iakoubov, R. & Brubaker, P. L. GPR119 is essential for oleoylethanolamide-induced glucagon-like peptide-1 secretion from the intestinal enteroendocrine L-cell. Diabetes 58, 1058–1066 (2009).
Yang, J. W., Kim, H. S., Choi, Y. W., Kim, Y. M. & Kang, K. W. Therapeutic application of GPR119 ligands in metabolic disorders. Diabetes, Obes. Metab. 20, 257–269 (2018).
Engelstoft, M. S. et al. Structural basis for constitutive activity and agonist-induced activation of the enteroendocrine fat sensor GPR119. Br. J. Pharmacol. 171, 5774–5789 (2014).
Bahirat, U. A., Shenoy, R. R., Goel, R. N. & Nemmani, K. V. APD668, a G protein-coupled receptor 119 agonist improves fat tolerance and attenuates fatty liver in high-trans fat diet induced steatohepatitis model in C57BL/6 mice. Eur. J. Pharmacol. 801, 35–45 (2017).
Bahirat, U. A., Talwar, R., Shenoy, R. R., Nemmani, K. V. & Goel, R. N. Combination of APD668, a G protein-coupled receptor 119 agonist with linagliptin, a DPPIV inhibitor, prevents progression of steatohepatitis in a murine model of non-alcoholic steatohepatitis with diabetes. Med. Mol. Morphol. 52, 36–43 (2019).
Rasmussen, S. G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Zhuang, Y. et al. Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Cell 184, 931–942.e18 (2021).
Fan, H. et al. Structural basis for ligand recognition of the human thromboxane A2 receptor. Nat. Chem. Biol. 15, 27–33 (2019).
Kumar, K. K. et al. Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176, 448–458 (2019).
Xing, C. et al. Cryo-EM structure of the human cannabinoid receptor CB2-Gi signaling complex. Cell 180, 645–654 e613 (2020).
Chrencik, J. E. et al. Crystal structure of antagonist bound human lysophosphatidic acid receptor. Cell 161, 1633–1643 (2015).
Hanson, M. A. et al. Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 (2012).
Cao, C. et al. Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat. Struct. Mol. Biol. 25, 488–495 (2018).
Kooistra, A. J. et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 49, D335–D343 (2021).
Xu, P. et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature https://doi.org/10.1038/s41586-021-03376-8 (2021).
Yuan, Y. et al. Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition. Cell Res. 31, 1263–1274 (2021).
Gusach, A. et al. Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors. Nat. Commun. 10, 5573 (2019).
Hori, T. et al. Na(+)-mimicking ligands stabilize the inactive state of leukotriene B4 receptor BLT1. Nat. Chem. Biol. 14, 262–269 (2018).
Michaelian, N. et al. Structural insights on ligand recognition at the human leukotriene B4 receptor 1. Nat. Commun. 12, 2971 (2021).
Srivastava, A. et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 (2014).
Liu, X. et al. Mechanism of beta2AR regulation by an intracellular positive allosteric modulator. Science 364, 1283–1287 (2019).
Zhuang, Y. et al. Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor. Cell Res. 31, 593–596 (2021).
Lu, J. et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 24, 570–577 (2017).
Yang, F. et al. Structural basis of GPBAR activation and bile acid recognition. Nature 587, 499–504 (2020).
Cherezov, V. et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007).
Garcia-Nafria, J., Nehme, R., Edwards, P. C. & Tate, C. G. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature https://doi.org/10.1038/s41586-018-0241-9 (2018).
Yin, W. et al. Crystal structure of the human 5-HT1B serotonin receptor bound to an inverse agonist. Cell Disco. 4, 12 (2018).
Wang, S. et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018).
Gaetani, S., Oveisi, F. & Piomelli, D. Modulation of meal pattern in the rat by the anorexic lipid mediator oleoylethanolamide. Neuropsychopharmacology 28, 1311–1316 (2003).
Lo Verme, J. et al. Regulation of food intake by oleoylethanolamide. Cell. Mol. Life Sci. 62, 708–716 (2005).
Astarita, G. et al. Postprandial increase of oleoylethanolamide mobilization in small intestine of the Burmese python (Python molurus). Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1412–R1412 (2006).
Zhang, S. Y. et al. Molecular matchmaking between the popular weight-loss herb Hoodia gordonii and GPR119, a potential drug target for metabolic disorder. Proc. Natl Acad. Sci. USA 111, 14571–14576 (2014).
Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976 (2012).
Liang, Y.-L. et al. Dominant Negative G Proteins Enhance Formation and Purification of Agonist-GPCR-G Protein Complexes for Structure Determination. ACS Pharmacol. Transl. Sci. 1(1), 12–20 (2018).
Xu, P. et al. Structures of the human dopamine D3 receptor-Gi complexes. Mol. Cell 81, 1147–1159. e1144 (2021).
Huang, S. et al. Structural basis for recognition of anti-migraine drug lasmiditan by the serotonin receptor 5-HT1F–G protein complex. Cell Res. 31, 1036–1038 (2021).
Mastronarde, D. N. J. J. O. S. B. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 152, 36–51 (2005).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. https://doi.org/10.1002/pro.3943 (2020).
Acknowledgements
The cryo-EM data were collected at the Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences (CAS) (Shanghai, China). This work was supported by the Ministry of Science and Technology (China) grant (2018YFA0507002 to H.E.X.); National Natural Science Foundation grants (32130022 to H.E.X., 82121005 to H.E.X., X.X. and Y.J., 81730099 to X.X., 32171187 to Y.J.); CAS Strategic Priority Research Program (XDB37030103 to H.E.X.); Shanghai Municipal Science and Technology Major Projects (2019SHZDZX02 to H.E.X. and TM202101H005 to X.X.), Shanghai Municipal Science and Technology Major Project (H.E.X.).
Author information
Authors and Affiliations
Contributions
P.X. and S.H. designed the expression constructs, purified the complexes, prepared samples for the cryo-EM as well as data collection and analysis. S.G., Y.Y. and Y.L. performed the functional assays and analyzed the data. X.C., X.H. and H.J. were responsible for the docking studies. P.C. and H.Z. were responsible for the mass spectrum studies. Y.J. participated in the supervision of P.X. and S.H., analyzed the structures and edited the manuscript. X.X. supervised the functional studies. H.E.X., X.X. and Y.J. conceived, designed and supervised the overall project. H.E.X., X.X., Y.J., P.X., S.H. and S.G. participated in data analysis and interpretation, and wrote the manuscript with inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Florian Ullrich, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM data processing of the LPC-GPR119-Gs-Nb35 structure.
a, Flowchart of cryo-EM data analysis of the LPC-GPR119-Gs-Nb35 complex. b, Representative cryo-EM image (scale bar, 50 nm) from 4,976 movies. c, Representative 2D averages show features of GPR119 TMD and Gs heterotrimer (scale bar, 5 nm). d, ‘Gold-standard’ Fourier shell correlation curves of the LPC- GPR119-Gs-Nb35 complex. e, Cryo-EM map and model of the LPC-GPR119-Gs-Nb35 complex. Cryo-EM density map and model are shown for all seven transmembrane α-helices from GPR119, LPC, Gαs α5 helices, and Gαs αN helices. Numerical data for graphs in b,c,d are available as source data.
Extended Data Fig. 2 Structural comparisons and functional data of the GPR119 activation by LPC.
a, Structural comparison of GPR119 with β2AR (PDB: 3SN6) and D1R (PDB: 7JVQ). b, Concentration-response of cAMP accumulation by the activation of GPR119 induced by LPC with different acyl tail. c, Mutagenesis data of LPC induced GPR119 activation. d, Relative expression of GPR119 mutants. Values are shown as the mean ± s.e.m. from at least three independent experiments performed in triplicate.
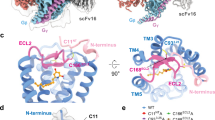
Extended Data Fig. 3 The unique TM5 structure of GPR119.
a-h, Structural comparisons of GPR119 with other class A GPCRs, including 5-HT1A (PDB: 7E2X), D1R (PDB: 7JVQ), CB1 (PDB: 6KPG), S1P1 (PDB: 7WF7), CLT1 (PDB: 6RZ4), BLT1 (PDB: 7K15), FFA1 (PDB: 5TZR), and LPA1 (PDB: 4Z34). i, The TM5 sequence alignment of GPR119 with other class A GPCRs shows a shift TM5 in GPR119 structure. The alignment was output from GPCRdb29 (gpcrdb.org) and edited based on the GPR119 structure.
Extended Data Fig. 4 Comparison of the orthosteric and allosteric (TM3-TM5-ICL3) pockets of GPR119 with other Class A GPCRs.
a, Structural comparison of the orthosteric ligand-binding pocket by GPR119 with other lipid-binding GPCRs, including CB1 (PDB: 6KPG), S1P1 (PDB: 7WF7), CLT1 (PDB: 6RZ4), BLT1 (PDB: 7K15), FFA1 (PDB: 5TZR), LPA1 (PDB: 4Z34), and PAFR (PDB: 5ZKP). b, Structural comparisons of the allosteric ligand-binding pocket (TM3-TM5-ICL3) by GPR119 with other class A GPCRs, including 5-HT1A (PDB: 7E2X), D1R (PDB: 7JVQ), CB1 (PDB: 6KPG), SIP1 (PDB: 7WF7), CLT1 (PDB: 6RZ4), BLT1 (PDB: 7K15), FFA1 (PDB: 5TZR), LPA1 (PDB: 4Z34). The allosteric binding pocket formed by TM3-TM5-ICL3 is circled with white dash lines.
Extended Data Fig. 5 Cryo-EM data processing of the APD668-GPR119-Gs-Nb35 structure.
a, Flowchart of cryo-EM data analysis of the APD668-GPR119-Gs-Nb35 complex. b, Representative cryo-EM image (scale bar, 50 nm) from 6,373 movies. c, Representative 2D averages show features of GPR119 TMD and Gs heterotrimer. d, ‘Gold-standard’ Fourier shell correlation curves of the APD668- GPR119-Gs-Nb35 complex. e, Cryo-EM map and model of the APD668-GPR119-Gs-Nb35 complex. Cryo-EM density map and model are shown for all seven transmembrane α-helices from GPR119, APD668, Gαs α5 helices, and Gαs αN helices.
Extended Data Fig. 6 Mutations increase the APD668 potency to GPR119.
a, d, g, cAMP assay shows mutations V85A (a), A89S (d), and A90S (g) increases the potency of APD668 to GPR119. Values are shown as the mean ± s.e.m. from at least three independent experiments performed in triplicate. b-c, the comparisons of the structure (b) and the V85A model (c) of APD668 bound-GPR119. e-f, the comparisons of the structure (e) and the A89S model (f) of APD668 bound-GPR119. h-i, the comparisons of the structure (h) and the A90S model (i) of APD668 bound-GPR119.
Extended Data Fig. 7 The activation of GPR119.
a-d, Structure of toggle switch residue W6.48 (a), PIF motif (b), DRY motif (c), and NPxxY motif (d) of GPR119 in compared with inactive and active β2AR, 5-HT1B, and D2R. e, The water molecule is coordinated by W2386.48, G2687.42, S2727.46, and N2717.45 of GPR119.
Extended Data Fig. 8 The unique interface of GPR119 ICL1 to the Gβ and Gαs subunits.
a-h, Structural comparisons at the ICL1 of GPR119-Gs with 5-HT1A-Gi, 5-HT1B-Gi, D1R-Gs, β2AR-Gs, CB1-Gi, and CB2-Gi complexes. i, Sequence alignment shows GPR119 has two more residues in the cytoplasmic end of the TM1 (1×61 and 1×62). The alignment was output from GPCRdb29 (gpcrdb.org) and edited based on the GPR119 structures.
Extended Data Fig. 9 Extended Data Fig.9 Docking and functional studies of fatty acid derivatives and a steroid glycoside Gordonoside F bind to GPR119.
a-d, Docking models of oleoylethanolamide (OEA, a), 2-oleoyl glycerol (2-OG, b), linoleylethanolamide (LEA, c), and 5-hydroxy-eicosapentaenoic acid (5-HEPE, d) to the GPR119 structure. e, Structure of Gordonoside F. f, The hydrophilic binding pocket is not favorable for Gordonoside F binding. g, The docking model of Gordonoside F binds to GPR119. h, Identification of the OEA-GPR119 recognition by cAMP assay. i, Identification of the Gordonoside F-GPR119 recognition by cAMP assay. Values are shown as the mean ± s.e.m. from at least three independent experiments performed in triplicate. UD indicates that the activation level is too low to determine EC50 values. Numerical data for graphs in h,i are available as source data.
Extended Data Fig. 10 Docking models of synthetic agonists to GPR119.
a, Comparison of the APD668-bound GPR119 structure with the docking models of 9 agonists to GPR119. b, The molecules used in docking studies and their docking scores.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, P., Huang, S., Guo, S. et al. Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119. Nat Struct Mol Biol 29, 863–870 (2022). https://doi.org/10.1038/s41594-022-00816-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00816-5
This article is cited by
-
Structural basis for lysophosphatidylserine recognition by GPR34
Nature Communications (2024)
-
Constitutive activation mechanism of a class C GPCR
Nature Structural & Molecular Biology (2024)
-
Structural genomics of the human dopamine receptor system
Cell Research (2023)
-
Specific binding of GPR174 by endogenous lysophosphatidylserine leads to high constitutive Gs signaling
Nature Communications (2023)
-
The activation mechanism and antibody binding mode for orphan GPR20
Cell Discovery (2023)