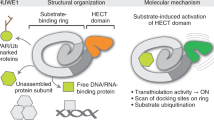
Abstract
The E2/E3 enzyme UBE2O ubiquitylates diverse clients to mediate important processes, including targeting unassembled ‘orphan’ proteins for quality control and clearing ribosomes during erythropoiesis. How quality-control factors, such as UBE2O, select clients on the basis of heterogeneous features is largely unknown. Here, we show that UBE2O client selection is regulated by ubiquitin binding and a cofactor, NAP1L1. Attaching a single ubiquitin onto a client enhances UBE2O binding and multi-mono-ubiquitylation. UBE2O also repurposes the histone chaperone NAP1L1 as an adapter to recruit a subset of clients. Cryo-EM structures of human UBE2O in complex with NAP1L1 reveal a malleable client recruitment interface that is autoinhibited by the intrinsically reactive UBC domain. Adding a ubiquitylated client identifies a distinct ubiquitin-binding SH3-like domain required for client selection. Our findings reveal how multivalency and a feed-forward mechanism drive the selection of protein quality-control clients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
EM maps and models are available under accession numbers EMD-26612, EMD-26614, EMD-26615, PDB 7UN3, and PDB 7UN6. All other data are available within the article and its Extended Data and Supplementary Information. Correspondence and requests for materials should be directed to S.S. Source data are provided with this paper.
References
Damgaard, R. B. The ubiquitin system: from cell signaling to disease biology and new therapeutic opportunities. Cell Death Differ. 28, 423–426 (2021).
Yanagitani, K., Juszkiewicz, S. & Hegde, R. S. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357, 472–475 (2017).
Nguyen, A. T. et al. UBE2O remodels the proteome during terminal erythroid differentiation. Science 357, eaan0218 (2017).
Berleth, E. S. & Pickart, C. M. Mechanism of ubiquitin conjugating enzyme E2-230K: catalysis involving a thiol relay? Biochemistry 35, 1664–1671 (1996).
Zhang, X. et al. Fine‐tuning BMP7 signalling in adipogenesis by UBE2O/E2‐230K‐mediated monoubiquitination of SMAD6. EMBO J. 32, 996–1007 (2013).
Mashtalir, N. et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell 54, 392–406 (2014).
Ullah, K., Zubia, E., Narayan, M., Yang, J. & Xu, G. Diverse roles of the E2/E3 hybrid enzyme UBE2O in the regulation of protein ubiquitination, cellular functions, and disease onset. FEBS J. 286, 2018–2034 (2019).
Chen, S. et al. Ubiquitin-conjugating enzyme UBE2O regulates cellular clock function by promoting the degradation of the transcription factor BMAL1. J. Biol. Chem. 293, 11296–11309 (2018).
Vila, I. K. et al. A UBE2O–AMPKα2 axis that promotes tumor initiation and progression offers opportunities for therapy. Cancer Cell 31, 208–224 (2017).
Huang, Y. et al. UBE2O targets Mxi1 for ubiquitination and degradation to promote lung cancer progression and radioresistance. Cell Death Differ. 28, 671–684 (2021).
Liu, X. et al. UBE2O promotes the proliferation, EMT and stemness properties of breast cancer cells through the UBE2O/AMPKα2/mTORC1-MYC positive feedback loop. Cell Death Dis. 11, 10 (2020).
Faust, T. B. et al. The HIV-1 Tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation. eLife 7, e31879 (2018).
Bagger, F. O., Kinalis, S. & Rapin, N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 47, D881–D885 (2019).
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011).
Wefes, I. et al. Induction of ubiquitin-conjugating enzymes during terminal erythroid differentiation. Proc. Natl Acad. Sci. USA 92, 4982–4986 (1995).
Brown, A., Baird, M. R., Yip, M. C., Murray, J. & Shao, S. Structures of translationally inactive mammalian ribosomes. eLife 7, e40486 (2018).
Husnjak, K. & Dikic, I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 (2012).
Huang, Q., Qin, D., Pei, D., Vermeulen, M. & Zhang, X. UBE2O and USP7 co‐regulate RECQL4 ubiquitinylation and homologous recombination‐mediated DNA repair. FASEB J. 36, e22112 (2022).
Zlatanova, J., Seebart, C. & Tomschik, M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 21, 1294–1310 (2007).
Park, Y.-J. & Luger, K. Structure and function of nucleosome assembly proteins. Biochem. Cell Biol. 84, 549–549 (2006).
Rodriguez, P. et al. Functional characterization of human nucleosome assembly Protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 44, 253–265 (1997).
Attia, M. et al. Interaction between nucleosome assembly protein 1-like family members. J. Mol. Biol. 407, 647–660 (2011).
Rössler, I. et al. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res. 47, 6984–7002 (2019).
Warren, C. & Shechter, D. Fly fishing for histones: catch and release by histone chaperone intrinsically disordered regions and acidic stretches. J. Mol. Biol. 429, 2401–2426 (2017).
Michelle, C., Vourc’h, P., Mignon, L. & Andres, C. R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 68, 616–628 (2009).
Bartke, T., Pohl, C., Pyrowolakis, G. & Jentsch, S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell 14, 801–811 (2004).
Sheng, Y. et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell Proteom. 11, 329–341 (2012).
Plechanovová, A., Jaffray, E., Tatham, M. H., Naismith, J. H. & Hay, R. T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Roldan, J. L. O. et al. Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications. PLoS One 8, e73018 (2013).
Stamenova, S. D. et al. Ubiquitin binds to and regulates a subset of SH3 domains. Mol. Cell 25, 273–284 (2007).
Elliott, P. R. et al. Regulation of CYLD activity and specificity by phosphorylation and ubiquitin-binding CAP-Gly domains. Cell Rep. 37, 109777 (2021).
Juszkiewicz, S. & Hegde, R. S. Quality control of orphaned proteins. Mol. Cell 71, 443–457 (2018).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Shimizu, Y. & Ueda, T. PURE technology. Methods Mol. Biol. 607, 11–21 (2010).
Feng, Q. & Shao, S. In vitro reconstitution of translational arrest pathways. Methods 137, 20–36 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Pettersen, E. F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213–221 (2010).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at biorXiv https://doi.org/10.1101/2021.10.04.463034 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. 74, 519–530 (2018).
Mirdita, M. et al. ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 66, 12–21 (2010).
Barad, B. A. et al. EMRinger: side chain–directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Schrödinger, L. & DeLano, W. The PyMOL Molecular Graphics System, Version 2.4 (2020).
Dong, R., Pan, S., Peng, Z., Zhang, Y. & Yang, J. mTM-align: a server for fast protein structure database search and multiple protein structure alignment. Nucleic Acids Res. 46, W380–W386 (2018).
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Acknowledgements
Cryo-EM data collection and screening were performed at the Harvard Center for Cryo-EM (HC2EM) and the Molecular Electron Microscopy Suite (MEMS) at Harvard Medical School. Data processing was supported by SBGrid. Mass spectrometry analysis was performed at the Taplin Mass Spectrometry Facility. The authors thank X. Wu and E. Goodall for experimental advice; A. Brown, D. Finley, T. Rapoport, W. Harper, and G. Nelson for critical reading; and S. Elsasser, M. Prado, and Shao lab members for useful discussions. This work was supported by NIH DP2GM137415, a Packard Fellowship, and the Vallee Foundation (S. S.), American Heart Association predoctoral fellowship 287375208 (M. C. J. Y), and NIH F31HL157976 (S. F. S.).
Author information
Authors and Affiliations
Contributions
M. C. J. Y. identified UBE2O ubiquitin binding activity and collected and processed cryo-EM data. S. F. S. identified NAP1L1. M. C. J. Y. and S. S. built atomic models. S. S. supervised the project. M. C. J. Y, S. F. S, and S. S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks Ivan Dikic and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor: Florian Ullrich, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 UBE2O has a ubiquitin-binding domain.
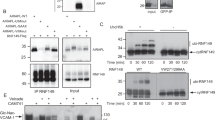
a, Expression (log2) values of UBE2O or IFRD2 in human hematopoietic cells at different stages of differentiation from hematopoietic stem cells (HSC) through the erythroid lineage13,14. Data (left to right) are from HSC (CD133 + CD34dim), HSC (CD38- CD34+), common myeloid progenitor (CMP) cells, megakaryocyte-erythroid progenitor (MEP) cells, and the following erythroid cell types: CD34 + CD71 + GlyA-, CD34- CD71 + GlyA-, CD34- CD71 + GlyA+, CD34- CD71lo GlyA + , CD34- CD71- GlyA +. b, Scheme of IFRD2 and ubiquitin-fused IFRD2 (Ub-IFRD2) variants analyzed in Fig. 1. c, SDS-PAGE and phosphorimaging of in vitro ubiquitylation timecourses, representative of 3 replicates, of radiolabeled uL2 fused to wildtype ubiquitin (Ub-uL2) or to ubiquitin with a mutated hydrophobic patch [Ub(3 A)-uL2], which were synthesized in the PURE translation system and then incubated with 500 nM UBE2O, 75 nM E1, 10 µM ubiquitin, and an energy regenerating system. Note: ubiquitylation (ubiq.) of Ub-uL2 is more efficient than that of Ub(3 A)-uL2. d, Autoradiography of in vitro ubiquitylation reactions of uL2 (top) and Ub-uL2 (bottom) as in c, with wildtype ubiquitin (WT Ub) or methylated ubiquitin (MeUb) incapable of polyubiquitin chain formation at the indicated timepoints. e, Coomassie staining of in vitro ubiquitylation reactions as in Fig. 1c of IFRD2 conjugated either to wildtype ubiquitin (Ub-IFRD2) or to ubiquitin in which all lysines are mutated to arginines [Ub(K0)-IFRD2] with WT Ub or MeUb. Note: the degree of client ubiquitylation in reactions shown in d and e does not change substantially with Ub(K0) or MeUb. Uncropped images for c-e and data for graphs in a are available as source data.
Extended Data Fig. 2 NAP1L1 interacts with UBE2O and UBE2O clients.
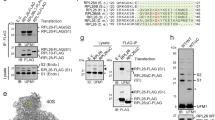
a, SDS-PAGE and SYPRO Ruby staining of pull-downs (PD) of a FLAG-tagged UBE2O client (uL14) or a FLAG-tagged non-UBE2O client (uS3) synthesized in a mammalian in vitro translation system. Labels indicate abundant bands which were excised and identified by mass spectrometry. Teal dotted boxes, UBE2O and UBE2O client. Purple dotted box indicates stoichiometric recovery of NAP1L1 with the UBE2O client but not the non-UBE2O client. b, UBE2O knockout Flp-In 293 T-REx cells co-expressing FLAG-tagged NAP1L1 without or with wildtype (WT) or catalytically dead (CD) Strep-tagged UBE2O were lysed (input), subjected to UBE2O PD, and analyzed by SDS-PAGE and immunoblotting, representative of >3 replicates. Note: NAP1L1 is not ubiquitylated and interacts equally well with WT and CD UBE2O. c, SDS-PAGE and Coomassie staining of sequential PDs of FLAG-tagged NAP1L1 and Strep-tagged UBE2O showing stoichiometric complex purification, representative of 2 replicates. FT, flow-through; Elu, elution. d, Cells co-expressing CD Strep-tagged UBE2O and a FLAG-tagged UBE2O client (uL14) were lysed and subjected to sequential UBE2O and client PDs. Input (in), FT, and Elu samples at the indicated relative concentrations were analyzed by SDS-PAGE and immunoblotting, representative of 2 replicates, suggesting recovery of ternary complexes of UBE2O, NAP1L1, and uL14. e, The radiolabeled UBE2O client uL14 was synthesized in vitro with either 1 µM untagged or FLAG-tagged NAP1L1 (F-NAP1L1), subjected to chemical crosslinking with 250 µM BMH as indicated, and analyzed directly (total) or after denaturing anti-FLAG immunoprecipitations (IP) by SDS-PAGE and autoradiography, representative of 2 replicates. Client crosslinks (x target) to UBE2O, NAP1L1, F-NAP1L1, and NAP1L1 or F-NAP1L1 dimers (x2) are indicated. Uncropped images for a-e are available as source data.
Extended Data Fig. 3 NAP1L1 recruits a subset of UBE2O clients.
a, Immunoblotting for UBE2O and NAP1 paralogs in wildtype (WT) or UBE2O knockout (ΔUBE2O) Flp-In 293 T-REx cells. b, Pull-down (PD) of WT Strep-tagged UBE2O co-expressed with HA-tagged uL2 or Ub-uL2 in UBE2O knockout (ΔU) or UBE2O/NAP1L1 double knockout (ΔUΔN) Flp-In 293 T-REx cells without or with induced re-expression of FLAG-tagged NAP1L1 (F-NAP1L1) incorporated into the Flp-In locus. F-NAP1L1 successfully rescues the interaction between uL2 and UBE2O which is impaired by knocking out NAP1L1, representative of 3 replicates. ubiq., ubiquitylated client. c, PD of WT or catalytically dead (CD) UBE2O co-expressed with FLAG-tagged IFRD2 in ΔUBE2O or ΔUBE2O ΔNAP1L1 Flp-In 293 T-REx cells, showing that knocking out NAP1L1 does not strongly impair the interaction between IFRD2 and UBE2O, representative of 2 replicates. d, UBE2O purifications from ΔU or ΔUΔN cells showing NAP1L1 and NAP1L4 association. e, PD of WT or CD UBE2O co-expressed with FLAG-tagged uL2 in ΔUBE2O, ΔUBE2O ΔNAP1L1, or UBE2O/NAP1L4 double knockout (ΔUBE2O ΔNAP1L4) Flp-In 293 T-REx cells. Note: knocking out NAP1L1 but not NAP1L4 impairs uL2 association with UBE2O, representative of 3 replicates. Uncropped images for a-e are available as source data.
Extended Data Fig. 4 Cryo-EM data processing of UBE2O in complex with NAP1L1.
a, SDS-PAGE and Coomassie staining of purified UBE2O-NAP1L1 complex without and with crosslinking with 250 µM BS3 used for cryo-EM analysis, representative of 2 independent preparations. b, Summary of processing strategy for the UBE2O + NAP1L1 dataset. Teal dotted box denotes class of apo UBE2O. Scale bar, 10 nm. c, Cryo-EM map of UBE2O-NAP1L1 colored by local resolution. Uncropped images for a are available as source data.
Extended Data Fig. 5 Cryo-EM maps and models.
a, Fourier shell correlation (FSC) coefficient vs. resolution (Å−1) curves of the indicated maps. Resolution was estimated at FSC = 0.143 (gray dotted line). b, Model vs. map FSC curves for structures as in a. c, Segmented EM map densities of the sharpened UBE2O-NAP1L1 map contoured at 8.6σ with corresponding atomic model. d-f, Secondary structure designations of the d, SH3-like domains, e, tandem SH3 domains, and f, UBC-CR3 of UBE2O colored by sequence conservation. Position of the catalytic cysteine is shown as a sphere. Connectivity between domains is indicated by lines with arrowheads and residue numbering. Dotted lines and labeled residues indicate interfaces involved in intra- and inter-molecular interactions. g, Net charge (calculated as +1 for K/R, + 0.5 for H, -1 for D/E) over sliding windows of 50 amino acids across the UBE2O sequence. Positions of CR1, CR2, and the beginning of UBC are indicated above. Red and blue circles indicate acidic and basic stretches, respectively.
Extended Data Fig. 6 Contributions to UBE2O client binding.
a, Model of UBE2O-NAP1L1 docked into a downsampled and unsharpened cryo-EM map contoured at 4.9σ showing the position of the β5-β6 hairpin loop of NAP1L1-B and the linear distance (dotted line) between the C-terminal end of SH3-C and the N-terminal end of the UBC domain on UBE2O, which are connected by residues 711-927. These features may influence client engagement. b, Position of UBC-charged ubiquitin (Ub, light orange) in the UBE2O-NAP1L1 complex relative to the putative client binding cavity, based on superposition of the UBE2O UBC domain with ubiquitin-charged UbcH5a (PDB 4AP4). Yellow sphere, position of C1040. Red asterisk denotes active site for client ubiquitylation. c, Angular distribution of a cryo-EM map of apo UBE2O. Preferential orientation was not improved despite combining three datasets of unmodified apo UBE2O and of PEGylated apo UBE2O without and with a stage tilt of 35°. d, Rigid body docking of the model of UBE2O in the UBE2O-NAP1L1 complex (Fig. 3c) into a cryo-EM map of apo UBE2O. Arrows indicate putative movements of the indicated domains to account for conformational differences.
Extended Data Fig. 7 Analysis of UBE2O E2 activity.
a, Superposition of UBC-CR3 of UBE2O (green) with the UBC domain (gray) of UbcH5a (PDB 2C4P, top), BIRC6 (PDB 3CEG, middle), or UBE2Z (PDB 5A4P, bottom). The E1 binding site (orange), the canonical RING E3 binding site (yellow), and the backside of the UBC domain (blue) are indicated relative to UbcH5a (top). Catalytic cysteines are shown as spheres. Note the additional helices (α5 and α6) present in UBC-CR3 of UBE2O and the UBC domains of BIRC6 and UBE2Z. b, In vitro ubiquitylation assays of a recombinant UBE2O client (Cmd1-DDX56)3 with full-length (FL) UBE2O, the CR1 and CR2 regions of UBE2O (UBE2O-N), and/or UBC-CR3 of UBE2O as indicated. The non-endogenous Cmd1-DDX56 client was analyzed due to its efficient ubiquitylation (ubiq.). Note: effective ubiquitylation is achieved with FL UBE2O or with both UBC-CR3 and UBEO-N, but not with either domain individually, representative of 2 replicates. c, Ubiquitin discharging assays from UBE2O UBC-CR3 (top) or UbcH5a (bottom) onto free lysine without or with a corresponding E3 activity (UBE2O-N or RNF4, respectively), representative of 3 independent experiments quantified in Fig. 4e. Ub~E2, ubiquitin-charged E2. Uncropped images for b and c are available as source data.
Extended Data Fig. 8 Cryo-EM data processing of UBE2O-NAP1L1 bound to Ub-uL2.
a, SDS-PAGE and Coomassie staining of purified Ub-uL2-UBE2O-NAP1L1 complex without and with crosslinking with 250 µM BS3 used for cryo-EM analysis, representative of 2 independent preparations. Note: Ub-uL2 is fused to UBE2O by a long linker but remains associated with the complex if this linker is cleaved after purification. b, Summary of processing strategy for the Ub-uL2-UBE2O + NAP1L1 dataset. Scale bar, 10 nm. c, Cryo-EM map of UBE2O-NAP1L1 bound to Ub-uL2 colored by local resolution. Uncropped images for a are available as source data.
Extended Data Fig. 9 Analysis of UBE2O-NAP1L1 with Ub-uL2.
a, Summary of processing strategy to examine conformational heterogeneity of UBE2O-NAP1L1 bound to ubiquitin. b, Superposition of models in which individual UBE2O and NAP1L1 domains were fitted to cryo-EM maps of the indicated classes (blue boxes) showing conformational heterogeneity in the relative positions and orientations of NAP1L1-B, UBC, and CR3 around the putative client binding interface. c, Models as in b, fitted to the indicated cryo-EM maps and superposed with the consensus model of the Ub-UBE2O-NAP1L1 complex (gray; Fig. 5b) colored by RMSD (blue – lowest, red – highest, normalized to each model). d, Processing strategy to generate subtracted particles of ubiquitin-bound UBE2O for focused refinement. e, Cryo-EM map resulting from focused refinement of ubiquitin-bound UBE2O as in d, colored by local resolution. f, Position of ubiquitin (orange) bound to SH3-C of UBE2O. Lysine at position 48 (K48) of ubiquitin is shown together with the distance (red) to the catalytic cysteine at position 1040 (yellow) of UBE2O. The C-terminal end of ubiquitin, which is modeled up to residue 73 is indicated by the orange dotted line leading to the putative client binding cavity.
Extended Data Fig. 10 SH3-C of UBE2O binds ubiquitin.
a, Superposition of ubiquitin (Ub; orange) bound to SH3-C of UBE2O (light blue) with the SH3 domain of Sla1 (top; PDB 2JT4) or the CAP-Gly SH3-like domain of CYLD (bottom; PDB 70WD) (light gray) bound to ubiquitin (dark gray). Note: although the SH3 and SH3-like domains are well-aligned, they bind ubiquitin through different interfaces. b, Pull-down (PD) of wildtype (WT), catalytically dead (CD), or I660A I662A (I660/662A or I-A) Strep-tagged UBE2O co-expressed with FLAG-tagged IFRD2, representative of 2 replicates. ubiq., ubiquitylated client. c, PD of WT or I660A I662A Strep-tagged UBE2O co-expressed with FLAG-tagged ubiquitin-fused uL2 (Ub-uL2) in UBE2O knockout (ΔUBE2O) or UBE2O/NAP1L1 double knockout (ΔUBE2O ΔNAP1L1) cells, representative of 2 replicates. Note: I660A I662A UBE2O loses ubiquitin-enhanced client binding activity. d, In vitro ubiquitylation timecourses of IFRD2 or ubiquitin-fused IFRD2 (Ub-IFRD2) with WT or I660A I662A UBE2O, representative of 2 replicates. Orange arrowheads, autoubiquitylated UBE2O. Uncropped images for b-d are available as source data.
Supplementary information
Supplementary Information
Supplementary Figures 1 and 2
Supplementary Video 1
Structure of UBE2O–NAP1L1 without and with ubiquitylated client
Supplementary Video 2
Conformational heterogeneity of UBE2O–NAP1L1 bound to ubiquitylated client
Supplementary Video 3
Interaction between UBE2O SH3-C and ubiquitin
Source data
Source Data Fig. 1
Unprocessed gels
Source Data Fig. 1c
Statistical source data
Source Data Fig. 2
Unprocessed gels
Source Data Fig. 2a
Size-exclusion chromatography data
Source Data Fig. 4
Unprocessed gels
SourceData Fig 4e
Statistical source data
Source Data Fig. 5
Unprocessed gels
Source Data Extended Data Fig. 1
Unprocessed gels
Source Data Extended Fig. 1a
Source data for transcript levels
Source Data Extended Data Fig. 2
Unprocessed gels
Source Data Extended Data Fig. 3
Unprocessed gels
Source Data Extended Data Fig. 4
Unprocessed gels
Source Data Extended Data Fig. 7
Unprocessed gels
Source Data Extended Data Fig. 8
Unprocessed gels
Source Data Extended Data Fig. 10
Unprocessed gels
Rights and permissions
About this article
Cite this article
Yip, M.C.J., Sedor, S.F. & Shao, S. Mechanism of client selection by the protein quality-control factor UBE2O. Nat Struct Mol Biol 29, 774–780 (2022). https://doi.org/10.1038/s41594-022-00807-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00807-6
This article is cited by
-
Noncanonical assembly, neddylation and chimeric cullin–RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex
Nature Structural & Molecular Biology (2024)