Abstract
Inosine-5′-monophosphate dehydrogenase (IMPDH), a key regulatory enzyme in purine nucleotide biosynthesis, dynamically assembles filaments in response to changes in metabolic demand. Humans have two isoforms: IMPDH2 filaments reduce sensitivity to feedback inhibition, while IMPDH1 assembly remains uncharacterized. IMPDH1 plays a unique role in retinal metabolism, and point mutants cause blindness. Here, in a series of cryogenic-electron microscopy structures we show that human IMPDH1 assembles polymorphic filaments with different assembly interfaces in extended and compressed states. Retina-specific splice variants introduce structural elements that reduce sensitivity to GTP inhibition, including stabilization of the extended filament form. Finally, we show that IMPDH1 disease mutations fall into two classes: one disrupts GTP regulation and the other has no effect on GTP regulation or filament assembly. These findings provide a foundation for understanding the role of IMPDH1 in retinal function and disease and demonstrate the diverse mechanisms by which metabolic enzyme filaments are allosterically regulated.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The coordinates are deposited in the PDB with accession codes PDB 7RER (interface-centered extended IMPDH1(514)), PDB 7RES (octamer-centered active IMPDH1(514)), PDB 7RFE (interface-centered compressed IMPDH1(514)), PDB 7RFG (octamer-centered compressed IMPDH1(514)), PDB 7RGL (interface-centered extended IMPDH1(546)), PDB 7RGM (octamer-centered extended IMPDH1(546)), PDB 7RGI (interface-centered compressed IMPDH1(546)), PDB 7RGQ (octamer-centered compressed IMPDH1(546)), PDB 7RFF (interface-centered extended IMPDH1(595)), PDB 7RFH (octamer-centered extended IMPDH1(595)), PDB 7RFI (interface-centered compressed IMPDH1(595)), PDB 7RGD (octamer-centered compressed IMPDH1(595). The cryo-EM maps are deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-24437 (interface-centered extended IMPDH1(514)), EMD-24438 (octamer-centered extended IMPDH1(514)), EMD-24439 (interface-centered compressed IMPDH1(514)), EMD-24441 (octamer-centered compressed IMPDH1(514)), EMD-24451 (interface-centered extended IMPDH1(546)), EMD-24452 (octamer-centered extended IMPDH1(546)), EMD-24450 (interface-centered compressed IMPDH1(546)), EMD-24454 (octamer-centered compressed IMPDH1(546)), EMD-24440 (interface-centered extended IMPDH1(595)), EMD-24442 (octamer-centered extended IMPDH1(595)), EMD-24443 (interface-centered compressed IMPDH1(595)) and EMD-24448 (octamer-centered compressed IMPDH1(595). Source data are provided with this paper.
References
Kennan, A. et al. On the role of IMPDH1 in retinal degeneration. Adv. Exp. Med. Biol. 533, 13–18 (2003).
Ji, Y., Gu, J., Makhov, A. M., Griffith, J. D. & Mitchell, B. S. Regulation of the interaction of inosine monophosphate dehydrogenase with mycophenolic acid by GTP. J. Biol. Chem. 281, 206–212 (2006).
Carcamo, W. C., Calise, S. J., von Mühlen, C. A., Satoh, M. & Chan, E. K. L. Molecular cell biology and immunobiology of mammalian rod/ring structures. Int. Rev. Cell Mol. Biol. 308, 35–74 (2014).
Calise, S. J. et al. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell. Mol. Life Sci. 71, 2963–2973 (2014).
Jackson, R. C., Weber, G. & Morris, H. P. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 256, 331–333 (1975).
Senda, M. & Natsumeda, Y. Tissue-differential expression of two distinct genes for human IMP dehydrogenase (E.C.1.1.1.205). Life Sci. 54, 1917–1926 (1994).
Carr, S. F., Papp, E., Wu, J. C. & Natsumeda, Y. Characterization of human type I and type II IMP dehydrogenases. J. Biol. Chem. 268, 27286–27290 (1993).
Calise, S. J., Abboud, G., Kasahara, H., Morel, L. & Chan, E. K. L. Immune response-dependent assembly of IMP dehydrogenase filaments. Front. Immunol. 9, 2789 (2018).
Duong-Ly, K. C. et al. T cell activation triggers reversible inosine-5′-monophosphate dehydrogenase assembly. J. Cell Sci. https://doi.org/10.1242/jcs.223289 (2018).
Johnson, M. C. & Kollman, J. M. Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation. eLife 9, e53243 (2020).
Fernández-Justel, D. et al. A nucleotide-dependent conformational switch controls the polymerization of Human IMP dehydrogenases to modulate their catalytic activity. J. Mol. Biol. 431, 956–969 (2019).
Hedstrom, L. IMP dehydrogenase-linked retinitis pigmentosa. Nucleosides Nucleotides Nucleic Acids 27, 839–849 (2008).
Zhang, R. et al. Characteristics and crystal structure of bacterial inosine-5’-monophosphate dehydrogenase. Biochemistry 38, 4691–4700 (1999).
Hedstrom, L. IMP dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109, 2903–2928 (2009).
Buey, R. M. et al. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun. 6, 8923 (2015).
Fernández-Justel, D., Peláez, R., Revuelta, J. L. & Buey, R. M. The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates. J. Biol. Chem. 294, 14768–14775 (2019).
Anthony, S. A. et al. Reconstituted IMPDH polymers accommodate both catalytically active and inactive conformations. Mol. Biol. Cell https://doi.org/10.1091/mbc.E17-04-0263 (2017).
Buey, R. M. et al. A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci. Rep. 7, 2648 (2017).
Kennan, A., Aherne, A. & Palfi, A. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(-/-) mice. Hum. Mol. Genet. 11, 547–557 (2002).
Bowne, S. J. et al. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum. Mol. Genet 11, 559–568 (2002).
Grover, S., Fishman, G. A. & Stone, E. M. A novel IMPDH1 mutation (Arg231Pro) in a family with a severe form of autosomal dominant retinitis pigmentosa. Ophthalmology 111, 1910–1916 (2004).
Bowne, S. J. et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 47, 34–42 (2006).
Labesse, G. et al. MgATP regulates allostery and fiber formation in IMPDHs. Structure 21, 975–985 (2013).
Thomas, E. C. et al. Different characteristics and nucleotide binding properties of inosine monophosphate dehydrogenase (IMPDH) isoforms. PLoS ONE 7, e51096 (2012).
Plana-Bonamaisó, A. et al. Post-translational regulation of retinal IMPDH1 in vivo to adjust GTP synthesis to illumination conditions. eLife 9, e56418 (2020).
Mortimer, S. E. et al. IMP dehydrogenase type 1 associates with polyribosomes translating rhodopsin mRNA. J. Biol. Chem. 283, 36354–36360 (2008).
Sullivan, L. S. et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest. Ophthalmol. Vis. Sci. 47, 3052–3064 (2006).
Mortimer, S. E. & Hedstrom, L. Autosomal dominant retinitis pigmentosa mutations in inosine 5′-monophosphate dehydrogenase type I disrupt nucleic acid binding. Biochem. J. 390, 41–47 (2005).
Aherne, A. et al. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum. Mol. Genet. 13, 641–650 (2004).
Bowne, S. J. et al. Why do mutations in the ubiquitously expressed housekeeping gene IMPDH1 cause retina-specific photoreceptor degeneration? Invest. Ophthalmol. Vis. Sci. 47, 3754–3765 (2006).
Wong-Riley, M. T. T. Energy metabolism of the visual system. Eye Brain 2, 99–116 (2010).
Country, M. W. Retinal metabolism: a comparative look at energetics in the retina. Brain Res. 1672, 50–57 (2017).
Arshavsky, V. Y. & Burns, M. E. Photoreceptor signaling: supporting vision across a wide range of light intensities. J. Biol. Chem. 287, 1620–1626 (2012).
Luo, D.-G., Xue, T. & Yau, K.-W. How vision begins: an odyssey. Proc. Natl Acad. Sci. USA 105, 9855–9862 (2008).
Palczewski, K. Chemistry and biology of the initial steps in vision: the Friedenwald lecture. Invest. Ophthalmol. Vis. Sci. 55, 6651–6672 (2014).
Du, J., An, J., Linton, J. D., Wang, Y. & Hurley, J. B. How excessive cGMP impacts metabolic proteins in retinas at the onset of degeneration. Adv. Exp. Med. Biol. 1074, 289–295 (2018).
Charish, J. cAMP and photoreceptor cell death in retinal degeneration. Adv. Exp. Med. Biol. 1185, 301–304 (2019).
Spellicy, C. J. et al. Characterization of retinal inosine monophosphate dehydrogenase 1 in several mammalian species. Mol. Vis. 13, 1866–1872 (2007).
Gunter, J. H. et al. Characterisation of inosine monophosphate dehydrogenase expression during retinal development: differences between variants and isoforms. Int. J. Biochem. Cell Biol. 40, 1716–1728 (2008).
Andashti, B., Yazdanparast, R., Barzegari, E. & Galehdari, H. The functional impact of the C/N-terminal extensions of the mouse retinal IMPDH1 isoforms: a kinetic evaluation. Mol. Cell. Biochem. 465, 155–164 (2020).
Andashti, B., Yazdanparast, R., Motahar, M., Barzegari, E. & Galehdari, H. Terminal peptide extensions augment the retinal IMPDH1 catalytic activity and attenuate the ATP-induced fibrillation events. Cell Biochem. Biophys. 79, 221–229 (2021).
Spellicy, C. J. et al. Characterization of retinal inosine monophosphate dehydrogenase 1 in several mammalian species. Mol. Vis. 13, 1866–1872 (2007).
Keppeke, G. D. et al. IMP/GTP balance modulates cytoophidium assembly and IMPDH activity. Cell Div. 13, 5 (2018).
Carcamo, W. C. et al. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS ONE 6, e29690 (2011).
Juda, P., Smigová, J., Kováčik, L., Bártová, E. & Raška, I. Ultrastructure of cytoplasmic and nuclear inosine-5’-monophosphate dehydrogenase 2 ‘rods and rings’ inclusions. J. Histochem. Cytochem. 62, 739–750 (2014).
Xu, D. et al. Retinal isoforms of inosine 5′-monophosphate dehydrogenase type 1 are poor nucleic acid binding proteins. Arch. Biochem. Biophys. 472, 100–104 (2008).
Lynch, E. M. & Kollman, J. M. Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat. Struct. Mol. Biol. 27, 42–48 (2020).
Chang, C., Keppeke, G. D., Sung, L. & Liu, J. Interfilament interaction between IMPDH and CTPS cytoophidia. FEBS J. 285, 3753–3768 (2018).
Zhao, H. et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J. Biol. Chem. 290, 6705–6713 (2015).
Hunkeler, M. et al. Structural basis for regulation of human acetyl-CoA carboxylase. Nature 558, 470–474 (2018).
Hayward, D. et al. ANKRD9 is a metabolically-controlled regulator of IMPDH2 abundance and macro-assembly. J. Biol. Chem. 294, 14454–14466 (2019).
Sharma, A. K. & Rohrer, B. Sustained elevation of intracellular cGMP causes oxidative stress triggering calpain-mediated apoptosis in photoreceptor degeneration. Curr. Eye Res. 32, 259–269 (2007).
Simonet, J. C., Burrell, A. L., Kollman, J. M. & Peterson, J. R. Freedom of assembly: metabolic enzymes come together. Mol. Biol. Cell 31, 1201–1205 (2020).
Lynch, E. M., Kollman, J. M. & Webb, B. A. Filament formation by metabolic enzymes—a new twist on regulation. Curr. Opin. Cell Biol. 66, 28–33 (2020).
Lynch, E. M. et al. Human CTP synthase filament structure reveals the active enzyme conformation. Nat. Struct. Mol. Biol. 24, 507–514 (2017).
Stoddard, P. R. et al. Polymerization in the actin ATPase clan regulates hexokinase activity in yeast. Science 367, 1039–1042 (2020).
Hunkeler, M. et al. Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution (Protein Data Bank, 2018); https://doi.org/10.2210/pdb6g2d/pdb
Mossessova, E. & Lima, C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000).
Volpe, D. A., Hamed, S. S. & Zhang, L. K. Use of different parameters and equations for calculation of IC50 values in efflux assays: potential sources of variability in IC50 determination. AAPS J. 16, 172–180 (2014).
Suloway, C. et al. Fully automated, sequential tilt-series acquisition with Leginon. J. Struct. Biol. 167, 11–18 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. GCTF: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A. Algorithmic advances in single particle Cryo-EM data processing using CryoSPARC. Microsc. Microanal. 26, 2322–2323 (2020).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D. Struct. Biol. 74, 519–530 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. in International Tables for Crystallography Vol. F, 539–547 (Wiley, 2012).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank the Arnold and Mabel Beckman Cryo-EM Center at the University of Washington for electron microscope use. We thank J. Calise for valuable feedback on the manuscript. This work was supported by the US National Institutes of Health (grant nos. R01GM118396 and R21EY031546 and S10OD032290 to J.M.K., R01GM-083025 to J.R.P., T32 CA-009035 to J.C.S. and F31EY030732 and T32GM008268 to A.L.B.), Universidad de Salamanca (Junta de Castilla y León fellowship to D.F.-J.), and Ministerio de Economía y Competitividad (grant no. BFU2016-79237-P to R.M.B.).
Author information
Authors and Affiliations
Contributions
A.L.B. performed kinetics assays, protein purification, cryo-EM data collection and image processing, and structure analysis. C.N. optimized kinetics experiment design. M.S. and M.C.J. developed cryo-EM processing approaches and processed cryo-EM data. J.C.S. designed and performed immunofluorescence experiments. D.F.-L. produced WT retinal protein used for cryo-EM. J.Q. optimized cryo-EM sample preparation. R.M.B. and J.R.P. conceptualized experiments. A.L.B. and J.M.K designed experiments, performed data analysis and interpretation and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information
Nature Structural and Molecular Biology thanks Carsten Sachse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
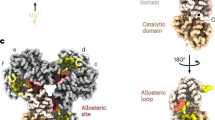
Extended Data Fig. 1 IMPDH structure and function.
a, Purine biosynthesis pathway. b, IMPDH monomer (6u9o) has a catalytic domain (green) that binds IMP and NAD+ in the active site, and a regulatory domain (pink) with three allosteric nucleotide binding sites. c, IMPDH is a tetramer in solution and can adopt a flat or bowed conformation. Side view of tetramers are depicted, so that only two monomers are visible. d, ATP (sites 1&2) or GTP (sites 2&3) binding promotes octamer assembly. e, IMPDH2 octamers can assemble into filaments of stacked octamers.
Extended Data Fig. 2 IMPDH1 Sequence Alignment.
Evolutionary conservation of the helix in the N-terminus of the longer retinal splice variant (blue) and the first 12 canonical residues particularly tyrosine 12 (pink).
Extended Data Fig. 3 IMPDH2-WT filament resists GTP inhibition.
a,b, GTP inhibition curves of IMPDH2 or IMPDH1-WT (solid line) and the respective non-assembly Y12A protein (dashed line). Individual data points are shown as diamonds (IMPDH2) or circles (IMPDH1), where filled are WT and empty Y12A. Reactions were performed in triplicate and the average for each concentration is shown as a bold rectangle (filled is WT, empty is Y12A). Error bars are standard deviation calculated from n = 3. Reactions performed with 1 µM protein, 1 mM ATP, 1 mM IMP, 300 µM NAD+, and varying GTP.
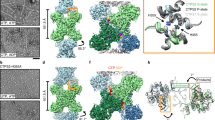
Extended Data Fig. 4 Cryo-EM workflow.
Flow chart summarizing data processing strategy for IMPDH1+ ATP/IMP/NAD+.
Extended Data Fig. 5 IMPDH1 active site map and model.
a-f, Cartoon representation of the active site. Side chains around the active site are shown as sticks. Chain A is dark green while the neighboring chain is light green. NAD+ is yellow and IMP red. Density for the ligand(s) is shown as a surface. a, IMPDH1(514) bound to ATP/IMP/NAD+. b, IMPDH1(514) bound to GTP/ATP/IMP. c, IMPDH1(546) bound to ATP/IMP/NAD+. d, IMPDH1(546) bound to GTP/ATP/IMP/NAD+. e, IMPDH1(595) bound to ATP. f, IMPDH1(595) bound to GTP/ATP/IMP/NAD+.
Extended Data Fig. 6 Inhibited IMPDH1-WT tetramer is in a bowed conformation.
a, Comparison of the catalytic tetramers of inhibited IMPDH2 filament (gray; 6u8s) to inhibited IMPDH2 free octamer (6uaj). Aligned on monomers with asterisk, other monomer pair has an alpha carbon RMSD of 2.1 Å. b, Comparison of the catalytic tetramers of inhibited IMPDH2 filament (gray; 6u8s) to inhibited IMPDH1 filament. Aligned on monomers with asterisk, other monomer pair has an alpha carbon RMSD of 3.7 Å.
Extended Data Fig. 7 Y12A non-assembly mutations prevents assembly in IMPDH1 variants.
Negative stain EM of purified human IMPDH1. Non-assembly mutation Y12A breaks both ATP- and GTP-dependent assembly. Scale bar 100 nm. Reactions performed with 1 µM protein, 1 mM ATP if used, 1 mM GTP if used.
Extended Data Fig. 8 IMPDH1 retinal variant (546) is similar to canonical IMPDH1.
a-c, Active IMPDH1(546) filament bound to ATP, IMP, NAD+. a, Low-pass filtered cryo-EM reconstruction b, Interface-focused cryo-EM reconstruction. 8 monomers are colored by catalytic domain (green) and regulatory domain (pink). c, View of the top of an octamer from inside the filament. The surface area buried by the octamer interface is in aqua with the indicated total buried surface area. (Surface representation of the atomic model at the assembly interface, with buried residues in cyan). d-f, Inhibited IMPDH1(546) filament bound to GTP, ATP, IMP, NAD+. d, Low-pass filtered cryo-EM reconstruction e, Interface-focused cryo-EM reconstruction. 8 monomers are colored by catalytic domain (green) and regulatory domain (pink). f, View of the top of an octamer from inside the filament. The surface area buried by the octamer interface is in aqua with the indicated total buried surface area. (Surface representation of the atomic model at the assembly interface, with buried residues in cyan).
Extended Data Fig. 9 IMPDH1 Retinal Variant C-term disrupts interactions.
a, Evolutionary conservation of the C-terminus in canonical IMPDH1 and both retinal splice variants. b, Surface representation of octamer side view. Dotted box indicates the region shown in c-d. c-d, Each chain is a different color green, C-term residues 510–512 in orange, and IMP in purple. c, inhibited canonical IMPDH1. d, Inhibited retinal variant IMPDH1(546).
Extended Data Fig. 10 IMPDH1 disease mutants have a variety of assembly phenotypes.
Negative stain EM of purified human IMPDH1. Scale bar 100 nm. Reactions performed with 1 µM protein, 1 mM ATP, 5 mM GTP, 3 mM IMP, 5 mM NAD+.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Supplementary Video 1.
Comparison between large and small interface IMPDH1 filaments. Morph comparison between large interface of ATP/IMP/NAD+ IMPDH1 filament to small interface in GTP/ATP/IMP IMPDH1 filament.
Supplementary Data 1
Supplementary data for Supplemental Table 1.
Supplementary Data 2
Supplementary data for Supplemental Table 4.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Burrell, A.L., Nie, C., Said, M. et al. IMPDH1 retinal variants control filament architecture to tune allosteric regulation. Nat Struct Mol Biol 29, 47–58 (2022). https://doi.org/10.1038/s41594-021-00706-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00706-2
This article is cited by
-
Filament formation drives catalysis by glutaminase enzymes important in cancer progression
Nature Communications (2024)
-
Human PRPS1 filaments stabilize allosteric sites to regulate activity
Nature Structural & Molecular Biology (2023)
-
Glioma cell membrane camouflaged cinobufotalin delivery system for combinatorial orthotopic glioblastoma therapy
Nano Research (2023)
-
Agglomeration: when folded proteins clump together
Biophysical Reviews (2023)