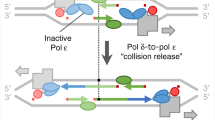
Abstract
Accurate DNA replication of an undamaged template depends on polymerase selectivity for matched nucleotides, exonucleolytic proofreading of mismatches, and removal of remaining mismatches via DNA mismatch repair (MMR). DNA polymerases (Pols) δ and ε have 3′–5′ exonucleases into which mismatches are partitioned for excision in cis (intrinsic proofreading). Here we provide strong evidence that Pol δ can extrinsically proofread mismatches made by itself and those made by Pol ε, independently of both Pol δ’s polymerization activity and MMR. Extrinsic proofreading across the genome is remarkably efficient. We report, with unprecedented accuracy, in vivo contributions of nucleotide selectivity, proofreading, and MMR to the fidelity of DNA replication in Saccharomyces cerevisiae. We show that extrinsic proofreading by Pol δ improves and balances the fidelity of the two DNA strands. Together, we depict a comprehensive picture of how nucleotide selectivity, proofreading, and MMR cooperate to achieve high and symmetrical fidelity on the two strands.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All whole genome sequencing data is available through Sequence Read Archive accession number PRJNA689775. Source data are provided with this paper.
Code availability
Muver suite is available via GitHub44 or upon request.
References
Lujan, S. A., Williams, J. S. & Kunkel, T. A. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 26, 640–654 (2016).
Kunkel, T. A. & Burgers, P. M. J. Arranging eukaryotic nuclear DNA polymerases for replication: Specific interactions with accessory proteins arrange Pols alpha, delta, and in the replisome for leading-strand and lagging-strand DNA replication. Bioessays https://doi.org/10.1002/bies.201700070 (2017).
Clausen, A. R. et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 22, 185–191 (2015).
Daigaku, Y. et al. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 22, 192–198 (2015).
Miyabe, I., Kunkel, T. A. & Carr, A. M. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 7, e1002407 (2011).
Nick McElhinny, S. A., Gordenin, D. A., Stith, C. M., Burgers, P. M. & Kunkel, T. A. Division of labor at the eukaryotic replication fork. Mol. Cell 30, 137–144 (2008).
Pursell, Z. F., Isoz, I., Lundstrom, E. B., Johansson, E. & Kunkel, T. A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317, 127–130 (2007).
Yeeles, J. T. P., Janska, A., Early, A. & Diffley, J. F. X. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell 65, 105–116 (2017).
Yu, C. et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell 56, 551–563 (2014).
Garbacz, M. A. et al. Evidence that DNA polymerase delta contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat. Commun. 9, 858 (2018).
Zhou, Z. X., Lujan, S. A., Burkholder, A. B., Garbacz, M. A. & Kunkel, T. A. Roles for DNA polymerase delta in initiating and terminating leading strand DNA replication. Nat. Commun. 10, 3992 (2019).
Aria, V. & Yeeles, J. T. P. Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. Mol. Cell. 73, 199–211.e10 (2018).
Guilliam, T. A. & Yeeles, J. T. P. Reconstitution of translesion synthesis reveals a mechanism of eukaryotic DNA replication restart. Nat. Struct. Mol. Biol. 27, 450–460 (2020).
Miyabe, I. et al. Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat. Struct. Mol. Biol. 22, 932–938 (2015).
Donnianni, R. A. et al. DNA polymerase delta synthesizes both strands during break-induced replication. Mol. Cell 76, 371–381 e4 (2019).
Bebenek, A. & Ziuzia-Graczyk, I. Fidelity of DNA replication-a matter of proofreading. Curr. Genet 64, 985–996 (2018).
Kunkel, T. A. Exonucleolytic proofreading. Cell 53, 837–840 (1988).
Li, G. M. Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008).
Barbari, S. R. & Shcherbakova, P. V. Replicative DNA polymerase defects in human cancers: Consequences, mechanisms, and implications for therapy. DNA Repair (Amst.) 56, 16–25 (2017).
Joyce, C. M. & Steitz, T. A. DNA polymerase I: from crystal structure to function via genetics. Trends Biochem. Sci. 12, 288–292 (1987).
Kornberg, T. & Kornberg, A. 4. Bacterial DNA Polymerases. in The Enzymes vol. 10 (ed. Boyer, P. D.) 119–144 (Academic Press, 1974).
Joyce, C. M. How DNA travels between the separate polymerase and 3′–5′-exonuclease sites of DNA polymerase I (Klenow fragment). J. Biol. Chem. 264, 10858–10866 (1989).
Nick McElhinny, S. A., Pavlov, Y. I. & Kunkel, T. A. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle 5, 958–962 (2006).
Bebenek, K., Matsuda, T., Masutani, C., Hanaoka, F. & Kunkel, T. A. Proofreading of DNA polymerase eta-dependent replication errors. J. Biol. Chem. 276, 2317–2320 (2001).
Perrino, F. W. & Loeb, L. A. Proofreading by the epsilon subunit of Escherichia coli DNA polymerase III increases the fidelity of calf thymus DNA polymerase alpha. Proc. Natl Acad. Sci. USA 86, 3085–3088 (1989).
Perrino, F. W. & Loeb, L. A. Hydrolysis of 3′-terminal mispairs in vitro by the 3′–5′ exonuclease of DNA polymerase delta permits subsequent extension by DNA polymerase alpha. Biochemistry 29, 5226–5231 (1990).
Pavlov, Y. I. et al. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 16, 202–207 (2006).
Morrison, A. & Sugino, A. The 3′→5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 242, 289–296 (1994).
St Charles, J. A., Liberti, S. E., Williams, J. S., Lujan, S. A. & Kunkel, T. A. Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair 31, 41–51 (2015).
Flood, C. L. et al. Replicative DNA polymerase delta but not epsilon proofreads errors in cis and in trans. PLoS Genet. 11, e1005049 (2015).
Bulock, C. R., Xing, X. & Shcherbakova, P. V. DNA polymerase delta proofreads errors made by DNA polymerase epsilon. Proc. Natl Acad. Sci. USA 117, 6035–6041 (2020).
Tran, H. T., Gordenin, D. A. & Resnick, M. A. The 3′→5′ exonucleases of DNA polymerases delta and epsilon and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 2000–2007 (1999).
Longley, M. J., Pierce, A. J. & Modrich, P. DNA polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem. 272, 10917–10921 (1997).
Kadyrov, F. A. et al. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl Acad. Sci. USA 106, 8495–8500 (2009).
Shcherbakova, P. V. et al. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 278, 43770–43780 (2003).
Williams, J. S. et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase varepsilon. DNA Repair 11, 649–656 (2012).
Jain, R. et al. Crystal structure of yeast DNA polymerase epsilon catalytic domain. PLoS ONE 9, e94835 (2014).
Hogg, M. et al. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat. Struct. Mol. Biol. 21, 49–55 (2014).
Dua, R., Levy, D. L. & Campbell, J. L. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274, 22283–22288 (1999).
Swan, M. K., Johnson, R. E., Prakash, L., Prakash, S. & Aggarwal, A. K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 16, 979–986 (2009).
Herr, A. J., Kennedy, S. R., Knowels, G. M., Schultz, E. M. & Preston, B. D. DNA replication error-induced extinction of diploid yeast. Genetics 196, 677–691 (2014).
Tracy, M. A. et al. Spontaneous polyploids and antimutators compete during the evolution of Saccharomyces cerevisiae mutator cells. Genetics 215, 959–974 (2020).
Lujan, S. A. et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 24, 1751–1764 (2014).
Burkholder, A. B. et al. Muver, a computational framework for accurately calling accumulated mutations. BMC Genomics 19, 345 (2018).
Reijns, M. A. M. et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015).
Fortune, J. M. et al. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 280, 29980–29987 (2005).
Lujan, S. A. et al. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 8, e1003016 (2012).
Andrianova, M. A., Bazykin, G. A., Nikolaev, S. I. & Seplyarskiy, V. B. Human mismatch repair system balances mutation rates between strands by removing more mismatches from the lagging strand. Genome Res. 27, 1336–1343 (2017).
Fijalkowska, I. J., Schaaper, R. M. & Jonczyk, P. DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMS Microbiol. Rev. 36, 1105–1121 (2012).
Drake, J. W., Charlesworth, B., Charlesworth, D. & Crow, J. F. Rates of spontaneous mutation. Genetics 148, 1667–1686 (1998).
Roche, H., Gietz, R. D. & Kunz, B. A. Specificity of the yeast rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics 137, 637–646 (1994).
Pavlov, Y. I., Shcherbakova, P. V. & Kunkel, T. A. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics 159, 47–64 (2001).
Kraszewska, J., Garbacz, M., Jonczyk, P., Fijalkowska, I. J. & Jaszczur, M. Defect of Dpb2p, a noncatalytic subunit of DNA polymerase varepsilon, promotes error prone replication of undamaged chromosomal DNA in Saccharomyces cerevisiae. Mutat. Res. 737, 34–42 (2012).
Garbacz, M. et al. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair 29, 23–35 (2015).
Garbacz, M. A. et al. The absence of the catalytic domains of Saccharomyces cerevisiae DNA polymerase strongly reduces DNA replication fidelity. Nucleic Acids Res. 47, 3986–3995 (2019).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Haradhvala, N. J. et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat. Commun. 9, 1746 (2018).
Nick McElhinny, S. A., Kissling, G. E. & Kunkel, T. A. Differential correction of lagging-strand replication errors made by DNA polymerases α and ∆. Proc. Natl Acad. Sci. USA 107, 21070–21075 (2010).
Li, H. & O’Donnell, M. E. The eukaryotic CMG helicase at the replication fork: emerging architecture reveals an unexpected mechanism. Bioessays https://doi.org/10.1002/bies.201700208 (2018).
Picher, A. J. et al. Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res. 34, 3259–3266 (2006).
Xing, X. et al. A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat. Commun. 10, 374 (2019).
Morrison, A., Johnson, A. L., Johnston, L. H. & Sugino, A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12, 1467–1473 (1993).
Simon, M., Giot, L. & Faye, G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 10, 2165–2170 (1991).
Albertson, T. M. et al. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl Acad. Sci. USA 106, 17101–17104 (2009).
Rayner, E. et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer 16, 71–81 (2016).
Maslowska, K. H., Makiela-Dzbenska, K., Mo, J. Y., Fijalkowska, I. J. & Schaaper, R. M. High-accuracy lagging-strand DNA replication mediated by DNA polymerase dissociation. Proc. Natl Acad. Sci. USA 115, 4212–4217 (2018).
Fukushima, S., Itaya, M., Kato, H., Ogasawara, N. & Yoshikawa, H. Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J. Bacteriol. 189, 8575–8583 (2007).
Dervyn, E. et al. Two essential DNA polymerases at the bacterial replication fork. Science 294, 1716–1719 (2001).
Sanders, G. M., Dallmann, H. G. & McHenry, C. S. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell 37, 273–281 (2010).
Bruck, I., Goodman, M. F. & O’Donnell, M. The essential C family DnaE polymerase is error-prone and efficient at lesion bypass. J. Biol. Chem. 278, 44361–44368 (2003).
Randall, J. R., Nye, T. M., Wozniak, K. J. & Simmons, L. A. RNase HIII is important for Okazaki fragment processing in Bacillus subtilis. J. Bacteriol. 201, e00686–18 (2019).
Paschalis, V. et al. Interactions of the Bacillus subtilis DnaE polymerase with replisomal proteins modulate its activity and fidelity. Open Biol. 7, 170146 (2017).
Kazlauskas, D., Krupovic, M., Guglielmini, J., Forterre, P. & Venclovas, C. Diversity and evolution of B-family DNA polymerases. Nucleic Acids Res. 48, 10142–10156 (2020).
Thomas, D. C. et al. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry 30, 11751–11759 (1991).
Schmitt, M. W., Matsumoto, Y. & Loeb, L. A. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie 91, 1163–1172 (2009).
Kunkel, T. A., Hamatake, R. K., Motto-Fox, J., Fitzgerald, M. P. & Sugino, A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol. Cell Biol. 9, 4447–4458 (1989).
Korona, D. A., Lecompte, K. G. & Pursell, Z. F. The high fidelity and unique error signature of human DNA polymerase epsilon. Nucleic Acids Res. 39, 1763–1773 (2011).
Arana, M. E., Seki, M., Wood, R. D., Rogozin, I. B. & Kunkel, T. A. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36, 3847–3856 (2008).
Wosika, V. et al. New families of single integration vectors and gene tagging plasmids for genetic manipulations in budding yeast. Mol. Genet Genomics 291, 2231–2240 (2016).
Morrison, A., Bell, J. B., Kunkel, T. A. & Sugino, A. Eukaryotic DNA polymerase amino acid sequence required for 3′–5′ exonuclease activity. Proc. Natl Acad. Sci. USA 88, 9473–9477 (1991).
Jin, Y. H. et al. The 3′→5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl Acad. Sci. USA 98, 5122–5127 (2001).
Kostriken, R. & Heffron, F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb. Symp. Quant. Biol. 49, 89–96 (1984).
Zhou, Z. X., Williams, J. S. & Kunkel, T. A. Studying ribonucleotide incorporation: strand-specific detection of ribonucleotides in the yeast genome and measuring ribonucleotide-induced mutagenesis. J. Vis. Exp. 58020 (2018).
Drake, J. W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl Acad. Sci. USA 88, 7160–7164 (1991).
Larrea, A. A. et al. Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl Acad. Sci. USA 107, 17674–17679 (2010).
Acknowledgements
We thank D. Gordenin and R. Schaaper for critical reading of and thoughtful comments on the manuscript. We thank P. Mieczkowski and others from the High Throughput Sequencing Facility of UNC Chapel Hill for performing Illumina sequencing. This study was supported by Project Z01 ES065070 to T.A.K from the Division of Intramural Research of the NIH, NIEHS.
Author information
Authors and Affiliations
Contributions
Z.-X.Z. and T.A.K. conceived the project. Z.-X.Z. performed most of the experiments. Z.-X.Z. and S.A.L. analyzed the genome-wide mutation data. A.B.B. performed mutation calling using the muver pipeline. J.A.S. contributed to mutation accumulation experiments. J.D. purified the Pol ε holoenzyme. C.F. and J.S.W. contributed to Supplementary Table 3.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Genetic interaction between pol2-M644G and pol3-exo-.
Reporter gene assays were performed similar to Fig. 1. n > =15 independent cultures were used for fluctuation analysis for each genotype.
Extended Data Fig. 2 pol3-x mutant does not support colony growth.
Tetrad dissection from a heterozygous pol3-x/POL3-WT diploid strain. Plate was incubated at °C for 5 days.
Supplementary information
Supplementary Information
Supplementary Note, Figures 1–7, Tables 1–9, and Supplementary Source Data.
Supplementary Data
Source data for Supplementary Table 3 and Supplementary Fig. 6.
Source data
Source Data Fig. 1
Individual data points from fluctuation analysis.
Source Data Fig. 2
Substitution mutation rates for each genomic isolate.
Source Data Fig. 3
Rates of individual substitution type for each isolate.
Source Data Fig. 3
Uncropped image of PAGE gel.
Source Data Fig. 4
Substitution mutation rates for each genomic isolate; individual data points from fluctuation analysis.
Source Data Extended Data Fig. 1
Individual data points from fluctuation analysis.
Rights and permissions
About this article
Cite this article
Zhou, ZX., Lujan, S.A., Burkholder, A.B. et al. How asymmetric DNA replication achieves symmetrical fidelity. Nat Struct Mol Biol 28, 1020–1028 (2021). https://doi.org/10.1038/s41594-021-00691-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00691-6
This article is cited by
-
Efficient replication of human nuclear DNA
Cell Research (2022)