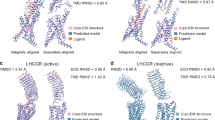
Abstract
We present an online, interactive platform for comparative analysis of all available G-protein coupled receptor (GPCR) structures while correlating to functional data. The comprehensive platform encompasses structure similarity, secondary structure, protein backbone packing and movement, residue–residue contact networks, amino acid properties and prospective design of experimental mutagenesis studies. This lets any researcher tap the potential of sophisticated structural analyses enabling a plethora of basic and applied receptor research studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data is available in GPCRdb (https://review.gpcrdb.org) and GitHub (https://github.com/protwis/gpcrdb_data). Documentation is available at https://docs.gpcrdb.org.
Code availability
All open-source code can be obtained from GitHub (https://github.com/protwis/protwis) under the permissive Apache 2.0 License (https://www.apache.org/licenses/LICENSE-2.0).
References
Foster, S. R. et al. Discovery of human signaling systems: pairing peptides to G protein-coupled receptors. Cell 179, 895–908 e21 (2019).
Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schioth, H. B. & Gloriam, D. E. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017).
Munk, C. et al. An online resource for GPCR structure determination and analysis. Nat. Methods 16, 151–162 (2019).
Zhou, Q. et al. Common activation mechanism of class A GPCRs. eLife 8, e50279 (2019).
Hauser, A. S. et al. GPCR activation mechanisms across classes and macro/microscales. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-021-00674-7(2021).
Venkatakrishnan, A. J. et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature 536, 484–487 (2016).
Flock, T. et al. Selectivity determinants of GPCR–G-protein binding. Nature 545, 317–322 (2017).
Sandhu, M. et al. Conformational plasticity of the intracellular cavity of GPCR–G-protein complexes leads to G-protein promiscuity and selectivity. Proc. Natl Acad. Sci. USA 116, 11956–11965 (2019).
Hauser, A. S. et al. Pharmacogenomics of GPCR drug targets. Cell 172, 41–54 e19 (2018).
Munk, C., Harpsoe, K., Hauser, A. S., Isberg, V. & Gloriam, D. E. Integrating structural and mutagenesis data to elucidate GPCR ligand binding. Curr. Opin. Pharmacol. 30, 51–58 (2016).
Kooistra, A. J. et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 49, D335–D343 (2021).
Kufareva, I. & Abagyan, R. Methods of protein structure comparison. Methods Mol. Biol. 857, 231–257 (2012).
Armstrong, D. R. et al. PDBe: improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 48, D335–D343 (2020).
Hilger, D. et al. Structural insights into differences in G protein activation by family A and family B GPCRs. Science 369, eaba3373 (2020).
Holm, L. & Sander, C. Mapping the protein universe. Science 273, 595–603 (1996).
Kayikci, M. et al. Visualization and analysis of non-covalent contacts using the Protein Contacts Atlas. Nat. Struct. Mol. Biol. 25, 185–194 (2018).
Venkatakrishnan, A. J. et al. Uncovering patterns of atomic interactions in static and dynamic structures of proteins. Preprint at bioRxiv https://doi.org/10.1101/840694 (2019).
Rousseeuw, P. J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Cock, P. J. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Rose, A. S. & Hildebrand, P. W. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 43, W576–W579 (2015).
Cao, C. et al. Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat. Struct. Mol. Biol. 25, 488–495 (2018).
Isberg, V. et al. Generic GPCR residue numbers—aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31 (2015).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Popov, P., Kozlovskii, I. & Katritch, V. Computational design for thermostabilization of GPCRs. Curr. Opin. Struct. Biol. 55, 25–33 (2019).
Muk, S. et al. Machine learning for prioritization of thermostabilizing mutations for G-protein coupled receptors. Biophys. J. 117, 2228–2239 (2019).
Bhattacharya, S., Lee, S., Grisshammer, R., Tate, C. G. & Vaidehi, N. Rapid computational prediction of thermostabilizing mutations for G protein-coupled receptors. J. Chem. Theory Comput. 10, 5149–5160 (2014).
Schonegge, A. M. et al. Evolutionary action and structural basis of the allosteric switch controlling beta2AR functional selectivity. Nat. Commun. 8, 2169 (2017).
Seifert, R. & Wenzel-Seifert, K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn-Schmiedeberg’s Arch. Pharmacol. 366, 381–416 (2002).
Waldhoer, M. et al. The carboxyl terminus of human cytomegalovirus-encoded 7 transmembrane receptor US28 camouflages agonism by mediating constitutive endocytosis. J. Biol. Chem. 278, 19473–19482 (2003).
Acknowledgements
This work was supported by the Lundbeck Foundation (grant nos R163-2013-16327 and R218-2016-1266), the Novo Nordisk Foundation (grant no NNF18OC0031226) and Independent Research Fund Denmark | Natural Sciences (grant no 8021-00173B) to D.E.G.
Author information
Authors and Affiliations
Contributions
D.E.G. conceptualized the study. D.E.G., C.M. and A.J.K. developed the methodology. A.J.K. curated the data. A.J.K. and D.E.G. carried out the investigation. A.J.K. and D.E.G. validated the data. D.E.G. wrote the original draft of the manuscript. A.J.K and A.S.H. reviewed and edited the manuscript. D.E.G., A.S.H. and A.J.K. visualized the study. D.E.G. acquired the funding. A.J.K. and C.M. developed the software. D.E.G. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
After the completion of this study, C.M. moved to become an employee of Novozymes A/S.
The other authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Peter Hildebrand and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Florian Ullrich was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 State-stabilizing mutation design tool, residue positions and experimental data.
a, The ‘State-stabilizing mutation design tool’ presents data-driven suggestions of mutagenesis experiments for all human GPCRs (https://review.gpcrdb.org/mutations/state_stabilizing). The tool ranks receptor positions by calculating a net sum of residue contacts expected to be gained or removed upon mutation. b, Suggested state-stabilizing positions for classes A and B1, respectively. These are limited to the 30 generic residue positions with the largest inactive/active state contact sum difference. The rightmost column indicates state stabilizers with high-frequency contacts5. c, Percent coverage of suggested state-stabilizing versus all other generic residue positions by experimentally determined mutations that are ligand activity-altering (>5-fold effect), thermostabilizing (540 data points) or expression increasing (100% would mean that all determinants or non-determinants, respectively are covered by experimental mutations). For ligand activity mutations (34,648 data points in GPCRdb), we required an effect in at least two receptors. For class A GPCRs, 27/30 residue positions have experimental support (avg. 1.8 functional associations). In class B1, 8 positions are supported by functional data (avg. 0.3 associations). We compared the percentages of residue positions covered by experimental effects for the class A and B1 determinants suggested in the mutation design tool (top 30) versus all other generic residue positions. This shows a near double representation of such data for suggested determinants than other generic residue positions in class B1. For class A GPCRs, we find stronger determinant overlaps spanning 2.1-, 2.9-, 3.1 and 8.8-fold ratios for mutations shown to influence thermostability, expression in ligand activity studies, ligand activity and expression of structure constructs, respectively. Notably, the top and third positions for class A GPCRs are two well-characterized residue microswitches, R3x50 and Y5x58 and the second position is a conserved proline causing the hinge of TM7. Notably, in both classes 13 out of 30 (43%) of suggested mutagenesis positions are unique from this tool (not in5.
Supplementary information
Rights and permissions
About this article
Cite this article
Kooistra, A.J., Munk, C., Hauser, A.S. et al. An online GPCR structure analysis platform. Nat Struct Mol Biol 28, 875–878 (2021). https://doi.org/10.1038/s41594-021-00675-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00675-6
This article is cited by
-
GLP-1R signaling neighborhoods associate with the susceptibility to adverse drug reactions of incretin mimetics
Nature Communications (2023)
-
Pathway selectivity in Frizzleds is achieved by conserved micro-switches defining pathway-determining, active conformations
Nature Communications (2023)
-
Recovery of the histamine H3 receptor activity lost in yeast cells through error-prone PCR and in vivo selection
Scientific Reports (2023)
-
GPCR activation mechanisms across classes and macro/microscales
Nature Structural & Molecular Biology (2021)