Abstract
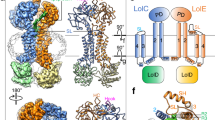
Lipoproteins in the outer membrane of Gram-negative bacteria are involved in various vital physiological activities, including multidrug resistance. Synthesized in the cytoplasm and matured in the inner membrane, lipoproteins must be transported to the outer membrane through the Lol pathway mediated by the ATP-binding cassette transporter LolCDE in the inner membrane via an unknown mechanism. Here, we report cryo-EM structures of Escherichia coli LolCDE in apo, lipoprotein-bound, LolA-bound, ADP-bound and AMP-PNP-bound states at a resolution of 3.2–3.8 Å, covering the complete lipoprotein transport cycle. Mutagenesis and in vivo viability assays verify features of the structures and reveal functional residues and structural characteristics of LolCDE. The results provide insights into the mechanisms of sorting and transport of outer-membrane lipoproteins and may guide the development of novel therapies against multidrug-resistant Gram-negative bacteria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Electron microscope density maps and atomic models have been deposited in the EMDB and PDB, respectively, with accession codes EMD-11882 and PDB 7ARH (lipoprotein-bound LolCDE), EMD-11887 and PDB 7ARM (lipoprotein-bound LolCDEA), EMD-11885 and PDB 7ARK (AMP-PNP-bound LolCDE with closed NBD), EMD-11884 and PDB 7ARJ (AMP-PNP-bound LolCDE with open NBD), EMD-11886 and PDB 7ARL (ADP-bound LolCDE), and EMD-11883 and PDB 7ARI (apo LolCDE). Source data are provided with this paper.
References
Fair, R. J. & Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 6, 25–64 (2014).
Raetz, C. R. H. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Sperandeo, P., Dehò, G. & Polissi, A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791, 594–602 (2009).
Dong, H., Tang, X., Zhang, Z. & Dong, C. Structural insight into lipopolysaccharide transport from the Gram-negative bacterial inner membrane to the outer membrane. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1461–1467 (2017).
Sperandeo, P., Martorana, A. M. & Polissi, A. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1451–1460 (2017).
Chimalakonda, G. et al. Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 108, 2492–2497 (2011).
Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014).
Bishop, R. E. Emerging roles for anionic non-bilayer phospholipids in fortifying the outer membrane permeability barrier. J. Bacteriol. 196, 3209–3213 (2014).
Raetz, C. R. H., Reynolds, C. M., Trent, M. S. & Bishop, R. E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 (2007).
Malinverni, J. C. & Silhavy, T. J. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl Acad. Sci. USA 106, 8009–8014 (2009).
Rowlett, V. W. et al. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199, e00849-16 (2017).
Bishop, R. E. et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram‐negative bacteria. EMBO J. 19, 5071–5080 (2000).
Isom, G. L. et al. LetB structure reveals a tunnel for lipid transport across the bacterial envelope. Cell 181, 653–664 (2020).
Ekiert, D. C. et al. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285 (2017).
Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 (2016).
Han, L. et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 192–196 (2016).
Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the β-barrel assembly machinery complex. Science 351, 180–186 (2016).
Iadanza, M. G. et al. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat. Commun. 7, 12865 (2016).
Tomasek, D. et al. Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583, 473–478 (2020).
Knowles, T. J., Scott-Tucker, A., Overduin, M. & Henderson, I. R. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 (2009).
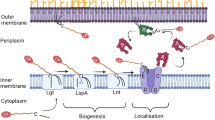
Narita, S., Matsuyama, S. & Tokuda, H. Lipoprotein trafficking in Escherichia coli. Arch. Microbiol. 182, 1–6 (2004).
Kovacs-Simon, A., Titball, R. W. & Michell, S. L. Lipoproteins of bacterial pathogens. Infect. Immun. 79, 548–561 (2011).
Tokuda, H. & Matsuyama, S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta Mol. Cell Res. 1693, 5–13 (2004).
Lorenz, C., Dougherty, T. J. & Lory, S. Correct sorting of lipoproteins into the inner and outer membranes of Pseudomonas aeruginosa by the Escherichia coli LolCDE transport system. Mbio 10, e00194-19 (2019).
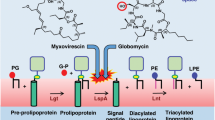
Yakushi, T., Masuda, K., Narita, S., Matsuyama, S. & Tokuda, H. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2, 212–218 (2000).
Narita, S. & Tokuda, H. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1414–1423 (2017).
Lorenz, C., Dougherty, T. J. & Lory, S. Transcriptional responses of Escherichia coli to a small-molecule inhibitor of LolCDE, an essential component of the lipoprotein transport pathway. J. Bacteriol. 198, 3162–3175 (2016).
McLeod, S. M. et al. Small-molecule inhibitors of Gram-negative lipoprotein trafficking discovered by phenotypic screening. J. Bacteriol. 197, 1075–1082 (2015).
Nickerson, N. N. et al. A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in Gram-negative bacteria. Antimicrob. Agents Chemother. 62, e02151-17 (2018).
Nayar, A. S. et al. Novel antibacterial targets and compounds revealed by a high-throughput cell wall reporter assay. J. Bacteriol. 197, 1726–1734 (2015).
Mori, H. & Ito, K. The Sec protein-translocation pathway. Trends Microbiol. 9, 494–500 (2001).
Mao, G. et al. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat. Commun. 7, 10198 (2016).
Vogeley, L. et al. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 351, 876–880 (2016).
Hillmann, F., Argentini, M. & Buddelmeijer, N. Kinetics and phospholipid specificity of apolipoprotein N-acyltransferase. J. Biol. Chem. 286, 27936–27946 (2011).
Narita, S. & Tokuda, H. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J. Biol. Chem. 282, 13372–13378 (2007).
Gennity, J. M. & Inouye, M. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 266, 16458–16464 (1991).
Seydel, A., Gounon, P. & Pugsley, A. P. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34, 810–821 (1999).
Masuda, K., Matsuyama, S. & Tokuda, H. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl Acad. Sci. USA 99, 7390–7395 (2002).
Okuda, S. & Tokuda, H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl Acad. Sci. USA 106, 5877–5882 (2009).
Kaplan, E., Greene, N. P., Crow, A. & Koronakis, V. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain. Proc. Natl Acad. Sci. USA 115, E7389–E7397 (2018).
Matsuyama, S., Yokota, N. & Tokuda, H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)‐dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 16, 6947–6955 (1997).
Takeda, K. et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 22, 3199–3209 (2003).
Tsukahara, J., Mukaiyama, K., Okuda, S., Narita, S. & Tokuda, H. Dissection of LolB function–lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 276, 4496–4504 (2009).
Nakada, S. et al. Structural investigation of the interaction between LolA and LolB using NMR. J. Biol. Chem. 284, 24634–24643 (2009).
Narita, S., Tanaka, K., Matsuyama, S. & Tokuda, H. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J. Bacteriol. 184, 1417–1422 (2002).
Taniguchi, N. & Tokuda, H. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J. Biol. Chem. 283, 8538–8544 (2008).
Tang, X. et al. Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nat. Struct. Mol. Biol. 28, 81–91 (2021).
Ito, Y., Kanamaru, K., Taniguchi, N., Shigehiko, M. & Tokuda, H. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol. Microbiol. 62, 1064–1075 (2006).
Okada, U. et al. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 8, 1336 (2017).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Tang, X. et al. Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat. Commun. 10, 4175 (2019).
Mizutani, M. et al. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex. FEBS Lett. 587, 23–29 (2013).
Crow, A., Greene, N. P., Kaplan, E. & Koronakis, V. Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl Acad. Sci. USA 114, 12572–12577 (2017).
Oguchi, Y. et al. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release. J. Biol. Chem. 283, 25414–25420 (2008).
Kanamaru, K., Taniguchi, N., Miyamoto, S., Narita, S. & Tokuda, H. Complete reconstitution of an ATP‐binding cassette transporter LolCDE complex from separately isolated subunits. FEBS J. 274, 3034–3043 (2007).
Rao, S. et al. Characterizing membrane association and periplasmic transfer of bacterial lipoproteins through molecular dynamics simulations. Structure 28, 475–487 (2020).
Khlebnikov, A., Risa, Ø., Skaug, T., Carrier, T. A. & Keasling, J. D. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 182, 7029–7034 (2000).
Liu, H. & Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. W. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189, 114–122 (2015).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank Y. Q. Wei and B. R. Dong for supporting the project and C. Ma for his help with protein purification. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0504803 and 2018YFA0507700 to X. Zhang) and the National Natural Science Foundation of China (31900039 to X.T., 32000844 to S.C. and 81971974 to H.D.), the Fundamental Research Funds for the Central Universities (2018XZZX001-13 to X. Zhang), the 1.3.5 Project for Disciplines Excellence of West China Hospital, Sichuan University (ZYYC20021) and Sichuan Science and Technology Program (2018TJPT0015 and 2018JY0094 to H.D).
Author information
Authors and Affiliations
Contributions
H.D. and X.T. conceived and designed the experiments. X.T., H.D. and Zhengyu Zhang made the constructs for protein expression. X.T., K.Z., Zhengyu Zhang, Q.L. and W.Q. expressed and purified the proteins. K.Z., Q.L., T.W., W.Q., C.S., Zhibo Zhang, X.W. and X. Zhu performed the mutagenesis, ATPase activities, the transport assays, cell-based assays and surface plasmon resonance analysis. X.T., H.D., K.Z., Q.L. and W.Q. prepared the samples. S.C., C.W. and X. Zhang undertook data collection, processing of electron microscopy data and structure constitution. H.D., X.T. and C.D. carried out model building and refinement. H.D. and X.T. wrote the manuscript and X. Zhang, S.C. and C.D. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Alessandra Polissi and Markus Seeger for their contribution to the peer review of this work. Florian Ullrich and Anke Sparmann were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Activity analysis of purified LolCDE complex.
a, Purified LolCDE was assessed for ATPase activity in detergent or liposomes at different ATP concentrations. b, c, The relative ATPase activities of LolCDE were measured in the presence of AMP-PNP (b) or ADP (c). d, In vivo cellular viability of the LolCDE silencing strain (HD200313) was studied. The LolCDE gene in HD200313 is controlled under an araBAD promotor that can only be induced in the presence of arabinose (method). Insertion of a leaky LolCDE expressing plasmid was used to test mutations in the LolCDE gene. e, ATPase activity determination of LolCDE with insertion of tags. f, LolCDE in liposomes of E. coli polar lipids were treated with or without proteinase K to assess the reconstitution orientations by SDS-PAGE. Data in d, f are representative results from n = 3 independent experiments. Data in a-c, e represent mean ± s.d. (n = 3 independent experiments). Statistics for panels a-c, e are available as Source Data. Uncropped images for panel d, f are available as Source Data.
Extended Data Fig. 2 Image-processing workflow for lipoprotein-bound LolCDE.
a, Cryo-EM microscope and selected two-dimensional class averages of cryo-EM particle images of lipoprotein-bound LolCDE. b, Scheme of three-dimensional classification, refinement of cryo-EM particle images, and the final 3D reconstitution of lipoprotein-bound LolCDE. c, Gold-standard FSC curves of the final cryo-EM maps of lipoprotein-bound LolCDE. d, The overall cryo-EM maps of lipoprotein-bound LolCDE are colored according to the local resolution. e, Cryo-EM maps with the atom model for individual transmembrane helices of LolC and LolE and the lipoprotein.
Extended Data Fig. 3 Image-processing workflow for AMP-PNP-bound LolCDE in closed and open NBD conformation.
a, Typical cryo-EM microscope and selected two-dimensional class averages of cryo-EM particle images of AMP-PNP-bound LolCDE. b, Scheme of three-dimensional classification, refinement of cryo-EM particle images and final 3D reconstitution of AMP-PNP-bound LolCDE with closed LolD at 4.1Å and open LolD at 3.2Å. c, Gold-standard FSC curves of the final cryo-EM maps of AMP-PNP-bound LolCDE.
Extended Data Fig. 4 Image-processing flowchart for apo LolCDE and lipoprotein bound-LolCDEA.
a, Typical cryo-EM microscope images of apo LolCDE and lipoprotein bound-LolCDEA. b, Selected two-dimensional class averages of cryo-EM particle images of apo LolCDE and lipoprotein bound-LolCDEA. c, Scheme of three-dimensional classification, refinement of cryo-EM particle images and the final 3D reconstitution of apo LolCDE at 3.4Å and lipoprotein bound-LolCDEA at 3.6Å. d, Gold-standard FSC curves of the final cryo-EM maps of apo LolCDE and lipoprotein bound LolCDEA .
Extended Data Fig. 5 Comparison of cryo-EM LolCDE structure to other known structures.
a, Lipoprotein-bound LolCDE (yellow) is superimposed to apo MacB (purple) (PDB code:5GKO). b, c, Dimerized conformations of LolCDE (orange) (b) and MacB (pink) (PDB 5LG7) (c) are superimposed to their apo state. d, PD of LolC is superimposed to crystal structure of LolC periplasmic domain (gray) (PDB code: 6F3Z). e, Superimposition between PDs of LolC (yellow) and LolE (green). f, Superimposition between PDs of LolC (yellow) and MacB (purple). The PD of MacB does not have the hook.
Extended Data Fig. 6 Topologic arrangement of LolC and LolE and surface representation of LolCDE.
a, Topology of LolC (yellow) showing the secondary structure arrangement. b, Topology of LolE (green) showing the secondary structure arrangement. c, Surface representation of lipoprotein-bound LolCDE. d, Cartoon representation of lipoprotein bound-LolCDE, rotated 180° along y-axis from c. e, Surface representation of apo LolCDE. f, Cartoon representation of apo LolCDE, rotated 90° along y-axis from e.
Extended Data Fig. 7 Conformational changes of LolCDE in different states.
a, Structural superimposition of apo LolCDE (cyan) to lipoprotein-bound LolCDE (yellow). b, Rotation of 90° from the left panel along y-axis. c, Structural superimposition of ADP-bound LolCDE (blue) to lipoprotein-bound LolCDE (yellow). d, Structural superimposition of AMP-PNP-bound LolCDE (un-dimerized LolD) (orange) to lipoprotein-bound LolCDE (yellow). e, Structural superimposition of AMP-PNP-bound LolD dimerized LolCDE (purple) to lipoprotein-bound LolCDE (yellow). f, Structural superimposition of four lipoprotein-bound structures, showing the same conformation regardless of nucleotide bindings.
Extended Data Fig. 8 Lipoprotein recognition in the central channel of LolCDE.
a, Residues of LolC and LolE showing interactions with the triacyl tails of the bound lipoprotein. Lipoprotein is shown in stick. b, Effects to lipoprotein binding upon mutations of lipoprotein peptide binding residues of LolC (F51D, L55D and D362A) and LolE (Y260E and R263A). c, Functional assays of mutants of lipoprotein binding residues. d, Protein leaky expression level control of the mutants. e, Periplasmic domains of LolC and LolE showing interactions of LolE with the peptide of the bound lipoprotein. f, Silver stain of purified LolCDE, showing a major band of E. coli lipoprotein Lpp along with protein LolCDE. g, MALDI-TOF/TOF/TOF mass spectrometry identification of Lpp peptide. Data in b-d, f are representative results from n = 3 independent experiments. Uncropped images for panels c, f are available as Source Data. Mass spectrometry raw data for g are available as Source Data.
Extended Data Fig. 9 Effect of nucleotide binding mutants of LolD.
a, b, Atomic interaction between the NBD of LolD and TMD of LolC (a) or LolE (b). c, Stabilization of the LolCDE complex was determined upon LolD mutations. d, Nucleotide (ADP) binding residues are shown at the interface of the NBDs of LolCDE. e, Size-exclusion chromatography of purified wild-type LolCDE and mutants of LolD. f, SDS-PAGE of purified LolCDE and mutants. g, ATPase activity was determined upon single mutations on LolD (K48A, E171A or S147E) in detergent or liposome. Data in c, e and f are representative results from n = 3 independent experiments. Data in g represent mean ± s.d. (n = 3 experiments). Statistics for panel g are available as Source Data.
Extended Data Fig. 10 Analysis of LolA binding to LolCDE.
a–d, SPR senorgram of lipoprotein-bound (a,c) or apo (b,d) LolA to lipoprotein-bound (a,b) or apo (c,d) LolCDE. e, Mean of KD values from three independent experiments with error bars ±SD, **p<0.01. f, Western-blot of analytes of LolA and LolCDE showing lipoprotein-bound or apo states. g, In the lipoprotein-bound LolCDEA structure, LolA docks on the lateral gate LolC-TM2/LolE-TM1 that is opposite to the lipoprotein-bound lateral gate LolC-TM1/LolE-TM2. h-i, Modelling LolA (light blue, h or wheat, i) in the AMP-PNP bound NBD-closed (h) or apo (i) LolCDE structures using the crystal structure of PD of LolC/LolA complex (PDB 6F3Z). j, Superimposition of lipoprotein-bound LolCDEA (red), modelled AMP-PNP bound LolCDEA (light blue) and modelled apo-LolCDEA (wheat) to compare the distinct positions of LolA related to LolCDE. Data in a-d are representative results from n = 3 independent experiments. Data in e represent mean ± s.d. (n = 3 independent experiments). Statistics for panels a-e are available as Source Data. Uncropped images for panel f are available as Source Data.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Source data
Source Data Fig. 1
Uncropped blots.
Source Data Fig. 3
Uncropped blots and agar plates.
Source Data Fig. 4
Uncropped blots and agar plates.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Uncropped blots and agar plates.
Source Data Extended Data Fig. 8
Mass spectrometry raw data.
Source Data Extended Data Fig. 8
Uncropped blots and agar plates.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Uncropped blots.
Source Data Extended Data Fig. 10
Statistical source data.
Source Data Extended Data Fig. 10
Uncropped blots.
Rights and permissions
About this article
Cite this article
Tang, X., Chang, S., Zhang, K. et al. Structural basis for bacterial lipoprotein relocation by the transporter LolCDE. Nat Struct Mol Biol 28, 347–355 (2021). https://doi.org/10.1038/s41594-021-00573-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00573-x
This article is cited by
-
Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii
Nature Chemical Biology (2023)
-
Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis
Nature Communications (2023)
-
Structural basis for triacylglyceride extraction from mycobacterial inner membrane by MFS transporter Rv1410
Nature Communications (2023)
-
A comparative analysis of lipoprotein transport proteins: LolA and LolB from Vibrio cholerae and LolA from Porphyromonas gingivalis
Scientific Reports (2023)
-
Envelope-Stress Sensing Mechanism of Rcs and Cpx Signaling Pathways in Gram-Negative Bacteria
Journal of Microbiology (2023)