Abstract
Suppressing cellular signal transducers of transcription 2 (STAT2) is a common strategy that viruses use to establish infections, yet the detailed mechanism remains elusive, owing to a lack of structural information about the viral–cellular complex involved. Here, we report the cryo-EM and crystal structures of human STAT2 (hSTAT2) in complex with the non-structural protein 5 (NS5) of Zika virus (ZIKV) and dengue virus (DENV), revealing two-pronged interactions between NS5 and hSTAT2. First, the NS5 methyltransferase and RNA-dependent RNA polymerase (RdRP) domains form a conserved interdomain cleft harboring the coiled-coil domain of hSTAT2, thus preventing association of hSTAT2 with interferon regulatory factor 9. Second, the NS5 RdRP domain also binds the amino-terminal domain of hSTAT2. Disruption of these ZIKV NS5-hSTAT2 interactions compromised NS5-mediated hSTAT2 degradation and interferon suppression, and viral infection under interferon-competent conditions. Taken together, these results clarify the mechanism underlying the functional antagonism of STAT2 by both ZIKV and DENV.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Coordinates and structure factors for the ZIKV RdRP–hSTAT21–713 complexes have been deposited in the Protein Data Bank with accession code PDB 6UX2. Cryo-EM density map and atomic coordinates of the NS5–hSTAT2 complex have been deposited to the EMDB and PDB under the accession codes EMD-21618 and PDB 6WCZ, respectively. Uncropped images and source data for the graphs are available in Supplementary Data 1 and 2, respectively.
References
Hoffmann, H. H., Schneider, W. M. & Rice, C. M. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 (2015).
Sadler, A. J. & Williams, B. R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008).
Chowdhury, F. Z. & Farrar, J. D. STAT2: a shape-shifting anti-viral super STAT. JAKSTAT 2, e23633 (2013).
Lim, C. P. & Cao, X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2, 536–550 (2006).
Shuai, K. et al. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76, 821–828 (1994).
Blaszczyk, K. et al. The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses. Cytokine Growth Factor Rev. 29, 71–81 (2016).
Fu, X. Y., Kessler, D. S., Veals, S. A., Levy, D. E. & Darnell, J. E. Jr ISGF3, the transcriptional activator induced by interferon α, consists of multiple interacting polypeptide chains. Proc. Natl Acad. Sci. USA 87, 8555–8559 (1990).
Platanitis, E. et al. A molecular switch from STAT2–IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 10, 2921 (2019).
Brierley, M. M. & Fish, E. N. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 25, 733–744 (2005).
Rasmussen, S. A., Jamieson, D. J., Honein, M. A. & Petersen, L. R. Zika virus and birth defects—reviewing the evidence for causality. N. Engl. J. Med. 374, 1981–1987 (2016).
Cao-Lormeau, V. M. et al. Guillain–Barre syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet 387, 1531–1539 (2016).
Gubler, D. J. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11, 480–496 (1998).
Ashour, J., Laurent-Rolle, M., Shi, P. Y. & Garcia-Sastre, A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83, 5408–5418 (2009).
Grant, A. et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19, 882–890 (2016).
Morrison, J. et al. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 9, e1003265 (2013).
Davidson, A. D. Chapter 2. New insights into flavivirus nonstructural protein 5. Adv. Virus Res. 74, 41–101 (2009).
Chen, X. et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93, 827–839 (1998).
Mao, X. et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol. Cell 17, 761–771 (2005).
Becker, S., Groner, B. & Muller, C. W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394, 145–151 (1998).
Li, J. et al. Structural basis for DNA recognition by STAT6. Proc. Natl Acad. Sci. USA 113, 13015–13020 (2016).
Neculai, D. et al. Structure of the unphosphorylated STAT5a dimer. J. Biol. Chem. 280, 40782–40787 (2005).
Duan, W. et al. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J. 36, 919–933 (2017).
Godoy, A. S. et al. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 8, 14764 (2017).
Wang, B. et al. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat. Commun. 8, 14763 (2017).
Zhao, B. et al. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 8, 14762 (2017).
Shuai, K., Stark, G. R., Kerr, I. M. & Darnell, J. E. Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-γ. Science 261, 1744–1746 (1993).
Improta, T. et al. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl Acad. Sci. USA 91, 4776–4780 (1994).
Uchil, P. D., Kumar, A. V. & Satchidanandam, V. Nuclear localization of flavivirus RNA synthesis in infected cells. J. Virol. 80, 5451–5464 (2006).
Martinez-Moczygemba, M., Gutch, M. J., French, D. L. & Reich, N. C. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-α-stimulated transcription factor ISGF3. J. Biol. Chem. 272, 20070–20076 (1997).
Rengachari, S. et al. Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc. Natl Acad. Sci. USA 115, E601–E609 (2018).
El Sahili, A. et al. NS5 from dengue virus serotype 2 can adopt a conformation analogous to that of its Zika virus and Japanese encephalitis virus homologues. J. Virol. 94, e01294–19 (2019).
Zhao, Y. et al. A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 11, e1004682 (2015).
Bussetta, C. & Choi, K. H. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry 51, 5921–5931 (2012).
Chaudhary, V. et al. Selective activation of type II interferon signaling by Zika virus NS5 protein. J. Virol. 91, e00163–17 (2017).
Kumar, A. et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 17, 1766–1775 (2016).
Speer, S. D. et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 7, 11496 (2016).
Xia, H. et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat. Commun. 9, 414 (2018).
Hertzog, J. et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur. J. Immunol. 48, 1120–1136 (2018).
Schwarz, M. C. et al. Rescue of the 1947 Zika virus prototype strain with a cytomegalovirus promoter-driven cDNA clone. Msphere 1, e00246–16 (2016).
Emeny, J. M. & Morgan, M. J. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43, 247–252 (1979).
Best, S. M. et al. Inhibition of interferon-stimulated JAK–STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79, 12828–12839 (2005).
Laurent-Rolle, M. et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK–STAT signaling. J. Virol. 84, 3503–3515 (2010).
Laurent-Rolle, M. et al. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16, 314–327 (2014).
Lin, R. J., Chang, B. L., Yu, H. P., Liao, C. L. & Lin, Y. L. Blocking of interferon-induced Jak–Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80, 5908–5918 (2006).
Lubick, K. J. et al. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe 18, 61–74 (2015).
Mazzon, M., Jones, M., Davidson, A., Chain, B. & Jacobs, M. Dengue virus NS5 inhibits interferon-α signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 200, 1261–1270 (2009).
Lu, G. & Gong, P. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 9, e1003549 (2013).
Upadhyay, A. K. et al. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr. F Struct. Biol. Commun. 73, 116–122 (2017).
Wang, B., Thurmond, S., Hai, R. & Song, J. Structure and function of Zika virus NS5 protein: perspectives for drug design. Cell. Mol. Life Sci. 75, 1723–1736 (2018).
Tay, M. Y. et al. The C-terminal 18 amino acid region of dengue virus NS5 regulates its subcellular localization and contains a conserved arginine residue essential for infectious virus production. PLoS Pathog. 12, e1005886 (2016).
Ng, I. H. W. et al. Zika virus NS5 forms supramolecular nuclear bodies that sequester importin-α and modulate the host immune and pro-inflammatory response in neuronal cells. ACS Infect. Dis. 5, 932–948 (2019).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 (1997).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Scheres, S. H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Vilas, J. L. et al. MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
This work was supported by a UC Cancer Research Coordinating Committee Faculty Seed Grant (CRR-20-634140) to J.S., and the NIH (1R35GM119721 to J.S, 1R21AI147057 to J.S. and R.H., R01GM071940 to Z.H.Z., R21 AI133649 to M.J.E. and R00GM111858 to S.E.O.). This work is also partly funded by the Trans fund of the State of California (AB2664) to J.S. and R.H, and by DHIPC (Dengue Human Immunology Project Consortium) and a NIAID funded program (U19AI118610) to A.G.-S. M.J.E. holds Investigator in Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund. K.Z. was partially supported by a fellowship from the China Scholarship Council. We acknowledge the assistance of D. Carter for microscopy imaging. Microscopy and/or image analysis was also performed at the Microscopy CoRE at the Icahn School of Medicine at Mount Sinai. We thank G. Tan for assisting with statistical analysis on the growth-curve data. We also acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057, 1S10OD018111 and U24GM116792) and NSF (DBI-1338135 and DMR-1548924). We would like to thank staff members at the Advanced Light Source, Lawrence Berkeley National Laboratory, for access to X-ray beamlines. The Advanced Light Source is supported by the director, office of science, office of basic energy sciences, of the US Department of Energy under contract no. DE-AC02-05CH11231. We thank S. Ding and D. Bogunovic for providing the DENV cDNA clone and VSV–GFP virus, respectively. We also thank Z. Hu for insightful discussions.
Author information
Authors and Affiliations
Contributions
B.W., S.T., K.Z., M.T.S.-A., J.F., J.L., L.G., W.R., Y.C., H.H., S.E.O., R.H. and J.S. performed experiments and data analysis. E.C.V. and M.J.E. provided technical support. R.H. and J.S. conceived the project, A.G.-S., Z.H.Z., R.H. and J.S. supervised the study. B.W., S.T., K.Z., Z.H.Z., R.H. and J.S. wrote the manuscript and all authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
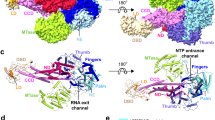
Extended Data Fig. 1 Structural analysis of the RdRP domain of ZIKV NS5 in complex with hSTAT21-713.
a, The structure of the NS5 RdRP-hSTAT21-713 complex, with hSTAT21-713 shown as electrostatic surface and RdRP shown as ribbon diagram. The two zinc clusters of NS5 RdRP are shown in expanded views, surrounded with Fo-Fc omit map (orange) contoured at 2.0 σ level. b, Two opposite views of the structure of the NS5 RdRP-hSTAT21-713 complex, with hSTAT21-713 shown as ribbon diagram and RdRP shown as electrostatic surface. The RNA entrance and exit tunnels of NS5 RdRP are marked with dashed circles (yellow). The disordered linkers of hSTAT21-713 are depicted as dashed lines. N- and C-termini are indicated by letter ‘N’ and ‘C’. c, d, Fo-Fc omit maps (violet) of selected residues for intermolecular interactions at the RdRP-CCD (c) and RdRP-ND (d) interfaces, contoured at 2.0 σ level. The color schemes for the RdRP and hSTAT2 are the same as that in Fig. 1.
Extended Data Fig. 2 Structural comparison of hSTAT2 and hSTAT1.
a, Ribbon representations of the CF of hSTAT2 (cyan) in association with its pY-tail segment (grey). The intramolecular interaction between the SH2 domain and the unphosphorylated pY-tail segment of hSTAT2 is highlighted in the expanded view. The hydrogen bonds are depicted as dashed lines. b, Ribbon representation of homodimeric hSTAT1 in complex with DNA, with the intermolecular interaction between the hSTAT1 SH2 domain (light pink) and the phosphorylated pY-tail segment from the other monomer (yellow) of the hSTAT1 homodimer shown in the expanded view. c, Sequence alignment of the pY-tail segments of human STAT proteins. Identical and similar residues are marked in red and blue, respectively. The target tyrosine is shaded in yellow. d, Structural model of the ZIKV NS5–hSTAT2–hSTAT1–DNA complex, with the surface electrostatic view of the hSTAT2 DBD shown in the expanded view.
Extended Data Fig. 3 Cryo-EM analysis of the ZIKV NS5–hSTAT2 complex.
a, A representative cryo-EM micrograph with select particles circled. b, 2D class averages used to generate the model. c, Local resolution evaluation of the reconstructed map by Resmap. d, e, Angular distribution (d) and global resolution evaluation based on ‘gold-standard’ Fourier shell correction (FSC) (e), both generated by RELION.
Extended Data Fig. 4 Details of the cryo-EM densities overlaid with their atomic models (ribbon) of the domains in ZIKV NS5–hSTAT2 complex.
a, Ribbon representations of the atomic model of NS5–hSTAT2, with NS5 (left) and hSTAT2 (right) overlaid with their respective density maps. b, c, Details for the MTase–CCD interaction, with densities of interacting residues shown as mesh. d, Details for the RdRP–CCD interaction, with densities of interacting residues shown as mesh. e, f, CCD, DBD and LD domains in hSTAT2. g–j, MTase, NE and Fingers, Thumb, and Palm domains in NS5.
Extended Data Fig. 5 Biochemical analysis of the interaction between ZIKV NS5 and hSTAT2.
a, Co-IP assay of 293T cells transfected with plasmids encoding C-terminally HA-tagged NS5 (NS5-HA) and FLAG-tagged hSTAT2 (hSTAT2-FLAG) or hSTAT1 (hSTAT1-FLAG). Immunoblot analysis of the whole cell extract (WCE) was used as input. IP was performed using antibodies against HA, FLAG and GAPDH. *antibody heavy chain. b, GST pull-down assay of the MTase and RdRP domains of ZIKV NS5 using GST-tagged full-length hSTAT2. c, Size-exclusion chromatography analysis of the interaction between hSTAT21-713 and ZIKV NS5. d, SDS-PAGE analysis of the elution fractions in (c). Uncropped gel images for a, b, d are shown in Supplementary Data 1.
Extended Data Fig. 6 Structural analysis of the ZIKV NS5–hSTAT2 interaction.
a, Superposition of the crystal structure of NS5 RdRP–hSTAT21-713 (grey) and the cryo-EM structure of full-length NS5–hSTAT21-713 (cyan). While the cryo-EM structure contains the MTase domain, there is no change in the relative orientations of different domains between the two structures. b, Structural superposition of free (PDB 5TMH) and hSTAT2-bound ZIKV NS5. The conformational transition of the MTase domain of ZIKV NS5 from free state to the hSTAT2-bound state is indicated by arrows. For clarity, all the hSTAT2 domains, except for the CCD, were removed. c, Surface view of the MTase domain of ZIKV NS5 bound to hSTAT2 CCD (green ribbon). The four helices of the CCD are sequentially numbered from ‘1’ to ‘4’. The potential RNA binding site is indicated by dashed circle. d, Surface of the DENV-3 MTase bound to viral cap-0 RNA (PDB 5DTO). e, hSTAT2 inhibition of ZIKV NS5–RNA binding assayed by electrophoretic mobility shift. Addition of NS5 to a cap-1 (m7GpppG2ʹOMe) yeast mRNA (YLR164W, 749 nt) resulted in a shift of the RNA band. Further addition of STAT2 resulted in release of RNA from the NS5–RNA complex, with concomitant accumulation of free RNA. Uncropped gel image for e is shown in Supplementary Data 1.
Extended Data Fig. 7 Mutational analysis of the interaction between ZIKV NS5 and hSTAT2.
a, IP analysis of endogenous hSTAT2 in 293T cells transfected with WT or mutYRDH NS5. Immunoblot was performed using antibodies against hSTAT2, HA, and GAPDH. b, In vitro pull-down assay of ZIKV NS5 mutations using GST-hSTAT2 as bait. wt: wild type. The experiment was repeated twice with consistent results. c, Quantification of nuclear localization of NS5, WT or mutants as shown in Fig. 3i. Data are mean ± s.d. (n = 3 biologically independent samples). Uncropped gel or blot images for a, b and source data for graphs in c are shown in Supplementary Data 1 and 2, respectively.
Extended Data Fig. 8 ZIKV NS5 competes against IRF9 for hSTAT2 binding.
a, Ribbon and surface view of the ZIKV NS5-binding sites (orange) of hSTAT2 CCD. b, Ribbon and surface view of the ZIKV NS5-binding sites (blue) of mouse STAT2 CCD. c, d, ITC binding curves of hSTAT2 over ZIKV NS5 (c) and IRF9 (d). The ITC binding assays were performed twice for the hSTAT2-NS5 interaction with similar results, and once for the hSTAT2-IRF9 interaction. e, Co-IP assay of HA-tagged ZIKV NS5 (NS5-HA), FLAG-tagged hSTAT2 (hSTAT2-FLAG) or hSTAT1 (hSTAT1-FLAG) and Myc-tagged IRF9 (IRF9-Myc). f, In vitro pull-down assay of ZIKV NS5 using GST-hSTAT2 in the presence of various amount of IRF9. g, IP assay of IRF9-Myc with hSTAT2-FLAG, wild type or mutant. The experiment in (g) was repeated once with similar results. Uncropped gel or blot images for e-g are shown in Supplementary Data 1.
Extended Data Fig. 9 Sequence and structural analyses of the DENV NS5–hSTAT2 interaction.
a–c, Sequence conservation analysis of the ZIKV NS5–hSTAT2 interaction using the ConSurf server (https://consurf.tau.ac.il/). The sequence conservation of hSTAT2 and ZIKV NS5 residues are colored coded and mapped onto the crystal structure of ZIKV RdRP–hSTAT2 (a) and cyro-EM structure of ZIKV NS5–hSTAT2 (b, c). d, Sequence alignment of the NS5 proteins from members of the Flaviviridae family, with the identical residues colored in red and the hSTAT2-binding sites of ZIKV NS5 marked by green asterisks. e, Representative negative-stain EM image of DENV NS5-hSTAT21-713. Bar: 40 nm. f, Characteristic 2D class averages with CTF correction. g, Structural overlay of DENV-3 NS5 and hSTAT2-bound ZIKV NS5. The potential steric clash between the ‘closed’ conformation of DENV-3 NS5 (PDB 4V0Q) and hSTAT2 is marked by a red circle.
Extended Data Fig. 10 Cellular analysis of the R327A (MutR) and G338E/D734A (MutGD) mutations of ZIKV NS5 on the hSTAT2 interaction and degradation.
a, Co-IP analysis showing the effects of MutR and MutGD in comparison with D734A (MutD) and MutYRDH on the NS5–hSTAT2 interaction. Immunoblot analysis of the IP and whole cell extract (WCE) was performed using antibodies against HA, FLAG and GAPDH. b, IF analysis of 293T cells transfected with plasmids encoding NS5-HA, wild type or mutant, and hSTAT2-FLAG. The transfected cells were fixed for immune staining using antibodies against HA (red) and FLAG (green). Nuclei were visualized by DAPI (blue) counter staining. c, Immunoblot analysis of 293T cells transfected with indicated plasmids encoding wild-type or mutant NS5-HAs using antibodies against HA, hSTAT2, and GAPDH, incubated with or without proteasome inhibitor MG132. d, Western-blot analysis of virus-infected A549 cells using antibodies against hSTAT2 and GAPDH. Uncropped blots for a, c, d are shown in Supplementary Data 1.
Supplementary information
Supplementary Information
Supplementary Figures 1 and 2.
Supplementary Video 1
Ribbon representation of the X-ray structure of ZIKV RdRP–hSTAT21–713 and zoomed-in view of the interface between RdRP and CCD.
Supplementary Video 2
Cryo-EM structure of ZIKV NS5–hSTAT21–713. The cryo-EM density map is shown as a shaded surface colored according to the domain color scheme in Fig. 1a. The atomic model derived from the cryo-EM map is first superposed with the cryo-EM density map and subsequently alone as a ribbon representation, followed by zoomed-in views of the interface between MTase and CCD.
Supplementary Data 1
Uncropped blots and gels
Supplementary Data 2
Statistical source data
Rights and permissions
About this article
Cite this article
Wang, B., Thurmond, S., Zhou, K. et al. Structural basis for STAT2 suppression by flavivirus NS5. Nat Struct Mol Biol 27, 875–885 (2020). https://doi.org/10.1038/s41594-020-0472-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0472-y
This article is cited by
-
A conformational selection mechanism of flavivirus NS5 for species-specific STAT2 inhibition
Communications Biology (2024)
-
Adaptation to host cell environment during experimental evolution of Zika virus
Communications Biology (2022)
-
Human Disease Phenotypes Associated with Loss and Gain of Function Mutations in STAT2: Viral Susceptibility and Type I Interferonopathy
Journal of Clinical Immunology (2021)