Abstract
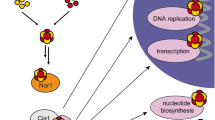
The cytosolic iron–sulfur (Fe–S) assembly (CIA) pathway is required for the insertion of Fe–S clusters into cytosolic and nuclear client proteins, including many DNA replication and repair factors. The molecular mechanisms of client protein recognition and Fe–S cluster transfer remain unknown. Here, we report crystal structures of the CIA targeting complex (CTC), revealing that its CIAO2B subunit is centrally located and bridges CIAO1 and the client adaptor protein MMS19. Cryo-EM reconstructions of human CTC bound either to the DNA replication factor primase or to the DNA helicase DNA2, combined with biochemical, biophysical and yeast complementation assays, reveal an evolutionarily conserved, bipartite client recognition mode facilitated by CIAO1 and the structural flexibility of the MMS19 subunit. Unexpectedly, the primase Fe–S cluster is located ~70 Å away from the CTC reactive cysteine, implicating conformational dynamics of the CTC or additional maturation factors in the mechanism of Fe–S cluster transfer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The atomic coordinates and structure factors reported in this study have been deposited in the Protein Data Bank (PDB) under accession codes 6TBN (CIAO1–CIAO2B complex), 6TBL (MMS19CTD–CIAO1–CIAO2B complex) and 6TC0 (MMS19–CIAO1–CIAO2B complex). The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-11016, EMD-11017 and EMD-11018 (CTC–primase), and EMD-11019, EMD-11020 and EMD-11021 (CTC–DNA2). Source data for Fig. 1f, Fig. 3, Extended Data Fig. 2b and Extended Data Fig. 6a are available with the paper online.
References
Beinert, H., Holm, R. H. & Munck, E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277, 653–659 (1997).
Netz, D. J. et al. Eukaryotic DNA polymerases require an iron–sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132 (2011).
Klinge, S., Hirst, J., Maman, J. D., Krude, T. & Pellegrini, L. An iron–sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 (2007).
Rudolf, J., Makrantoni, V., Ingledew, W. J., Stark, M. J. & White, M. F. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23, 801–808 (2006).
Landry, A. P. & Ding, H. The N-terminal domain of human DNA helicase Rtel1 contains a redox active iron-sulfur cluster. BioMed Res. Int. 2014, 285791 (2014).
Yeeles, J. T., Cammack, R. & Dillingham, M. S. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem. 284, 7746–7755 (2009).
Pokharel, S. & Campbell, J. L. Cross talk between the nuclease and helicase activities of Dna2: role of an essential iron–sulfur cluster domain. Nucleic Acids Res. 40, 7821–7830 (2012).
Bharti, S. K. et al. Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell. Mol. Life Sci. 71, 2625–2639 (2014).
Fan, L. et al. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 133, 789–800 (2008).
Gari, K. et al. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337, 243–245 (2012).
Fuss, J. O., Tsai, C. L., Ishida, J. P. & Tainer, J. A. Emerging critical roles of Fe–S clusters in DNA replication and repair. Biochim. Biophys. Acta 1853, 1253–1271 (2015).
Paul, V. D. & Lill, R. Biogenesis of cytosolic and nuclear iron–sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 1853, 1528–1539 (2015).
Lill, R. et al. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron–sulfur proteins. Eur. J. Cell Biol. 94, 280–291 (2015).
Kispal, G., Csere, P., Prohl, C. & Lill, R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989 (1999).
Netz, D. J. et al. A bridging [4Fe–4S] cluster and nucleotide binding are essential for function of the Cfd1–Nbp35 complex as a scaffold in iron–sulfur protein maturation. J. Biol. Chem. 287, 12365–12378 (2012).
Pallesen, L. J., Solodovnikova, N., Sharma, A. K. & Walden, W. E. Interaction with Cfd1 increases the kinetic lability of FeS on the Nbp35 scaffold. J. Biol. Chem. 288, 23358–23367 (2013).
Roy, A., Solodovnikova, N., Nicholson, T., Antholine, W. & Walden, W. E. A novel eukaryotic factor for cytosolic Fe–S cluster assembly. EMBO J. 22, 4826–4835 (2003).
Hausmann, A. et al. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron–sulfur protein assembly machinery. Proc. Natl Acad. Sci. USA 102, 3266–3271 (2005).
Balk, J., Pierik, A. J., Netz, D. J., Mühlenhoff, U. & Lill, R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulphur proteins. EMBO J. 23, 2105–2115 (2004).
Song, D. & Lee, F. S. A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 283, 9231–9238 (2008).
Odermatt, D. C. & Gari, K. The CIA targeting complex is highly regulated and provides two distinct binding sites for client iron-sulfur proteins. Cell Rep. 18, 1434–1443 (2017).
Prakash, L. & Prakash, S. Three additional genes involved in pyrimidine dimer removal in Saccharomyces cerevisiae: RAD7, RAD14 and MMS19. Mol. Gen. Genet. 176, 351–359 (1979).
Lauder, S. et al. Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol. Cell. Biol. 16, 6783–6793 (1996).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010).
Ouyang, B. et al. Solution structure of monomeric human FAM96A. J. Biomol. NMR 56, 387–392 (2013).
Srinivasan, V. et al. Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 15, 1246–1257 (2007).
Xu, C. & Min, J. Structure and function of WD40 domain proteins. Protein Cell 2, 202–214 (2011).
Stehling, O. et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195–199 (2012).
Maione, V., Cantini, F., Severi, M. & Banci, L. Investigating the role of the human CIA2A-CIAO1 complex in the maturation of aconitase. Biochim. Biophys. Acta Gen. Subj. 1862, 1980–1987 (2018).
Weon, J. L., Yang, S. W. & Potts, P. R. Cytosolic iron-sulfur assembly is evolutionarily tuned by a cancer-amplified ubiquitin ligase. Mol. Cell 69, 113–125.e6 (2018).
Vashisht, A. A., Yu, C. C., Sharma, T., Ro, K. & Wohlschlegel, J. A. The association of the xeroderma pigmentosum group D DNA helicase (XPD) with transcription factor IIH is regulated by the cytosolic iron-sulfur cluster assembly pathway. J. Biol. Chem. 290, 14218–14225 (2015).
Xu, Y. et al. Structure of the protein phosphatase 2A holoenzyme. Cell 127, 1239–1251 (2006).
Chook, Y. M. & Blobel, G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 (2001).
Cavadini, S. et al. Cullin–RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531, 598–603 (2016).
Fischer, E. S. et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 (2011).
Cook, A. G. & Conti, E. Nuclear export complexes in the frame. Curr. Opin. Struct. Biol. 20, 247–252 (2010).
Baek, K. et al. NEDD8 nucleates a multivalent cullin–RING–UBE2D ubiquitin ligation assembly. Nature 578, 461–466 (2020).
Paul, V. D. et al. The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion. Eife 4, e08231 (2015).
Abdulrahman, W. et al. A set of baculovirus transfer vectors for screening of affinity tags and parallel expression strategies. Anal. Biochem. 385, 383–385 (2009).
Zhao, Y., Chapman, D. A. & Jones, I. M. Improving baculovirus recombination. Nucleic Acids Res. 31, E6–E6 (2003).
Holt, M. E., Salay, L. E. & Chazin, W. J. A polymerase with potential: the Fe-S cluster in human DNA primase. Meth. Enzymol. 595, 361–390 (2017).
O’Brien, E. et al. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355, eaag1789 (2017).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Uson, I. & Sheldrick, G. M. An introduction to experimental phasing of macromolecules illustrated by SHELX; new autotracing features. Acta Crystallogr. D Struct. Biol. 74, 106–116 (2018).
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 (2003).
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010).
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Schröder, G. F., Levitt, M. & Brunger, A. T. Deformable elastic network refinement for low-resolution macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 70, 2241–2255 (2014).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68, 368–380 (2012).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Hohn, M. et al. SPARX, a new environment for cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Schenk, A. D., Cavadini, S., Thomä, N. H. & Genoud, C. Live analysis and reconstruction of single-particle cryo-electron microscopy data with CryoFLARE. J. Chem. Inf. Model. 60, 2561–2569 (2020).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank S. Cavadini and A. Schenk for help with cryo-EM data collection and processing and for advice about sample optimization. We are particularly grateful to the FMI core facilities: A. Graff Meyer and C. Genoud (electron microscopy); H. Gut, J. Keusch and G. Kempf (X-ray crystallography); and V. Iesmantavicius, D. Hess and J. Seebacher (mass spectrometry). Part of this work was performed at beamlines X06DA and X10SA of the Swiss Light Source (Paul Scherrer Institute). We thank the staff at both beamlines for assistance with X-ray data collection; W. Chazin (Vanderbilt University) for providing the plasmids for recombinant expression of human primase in bacteria and purified primase protein for initial studies; A. Potenza for help with protein expression in the early stage of the project; R. Bunker for help and advice with crystallographic data collection and processing; K. Shimada, M. Hauer and I. Deshpande for sharing protocols and advice on yeast experiments; K. Gari and D. Odermatt for sharing plasmids and helpful discussions; M. Jinek for advice on crystallographic data collection, processing and model building, and critical reading of the manuscript; and J. Reinert and F. Bleichert for comments on the manuscript. S.A.K. was supported by a long-term postdoctoral fellowship from the European Molecular Biology Organization (EMBO, ALTF 871–2014). This work was funded by the Swiss National Science Foundation through Sinergia grant number CRSII3_160734 and by the European Research Council under the European Union’s Horizon 2020 Research and Innovation program, grant number 666068, to N.H.T.
Author information
Authors and Affiliations
Contributions
Conceptualization was by S.A.K. and N.H.T. S.A.K. performed and devised the data curation, formal analysis, investigation, visualization and methodology. Project administration was by S.A.K. and N.H.T. The project was supervised by N.H.T. Validation and writing was carried out by S.A.K. and N.H.T.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Crystal structure of the CIAO1-CIAO2B core complex.
a, Coomassie-stained SDS-PAGE of purified Drosophila melanogaster CIAO1-CIAO2B complex. b, Surface representation of CIAO1 colored according to amino acid conservation across species based on the alignment in e. c, Surface representation of CIAO1 colored according to electrostatic potential from -3 to +3 kBT/e. d, Strep pull-down assay from Hi5 insect cells expressing Strep-CIAO2B and His-CIAO1 wt or indicated mutants. e, CIAO1 sequence alignment used to map evolutionary conservation in b. Secondary structure elements of human CIAO1 are indicated above the alignment. Numbering is relative to the human sequence. f, Yeast drop assay of CIAO1 point mutations located at the top face of the β-propeller. A Gal-CIA1 strain was transformed with either empty vector, wt human CIAO1, or a CIAO1 point mutant; a dilution series of each culture was spotted on agar plates in the presence of glucose or galactose.
Extended Data Fig. 2 Crystal structures of the MMS19-CIAO2B-CIAO1 CIA targeting complex.
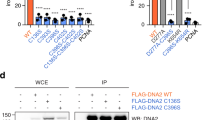
a, Coomassie-stained SDS-PAGE gels of purified MMS19CTD-CIAO2B-CIAO1 and MMS19-CIAO2B-CIAO1 complexes. b, Pull-down assay from HEK293 cells using Flag-tagged human MMS19 wt and mutants, and myc-tagged human CIAO2B/CIAO1. WCE (right) and eluted proteins (IP:Flag, left) were analyzed by Western blot. Corresponding to Fig. 1f. c, Yeast drop assay of MMS19 mutations. A MMS19 knockout strain was transformed with either empty galactose-inducible p415GAL1 vector, p415GAL1 expressing wt MMS19, or p415GAL1 vectors expressing MMS19 mutants. A dilution series of each culture was spotted on agar plates in the presence or absence of galactose and 20 mM HU. d, Lysine residues implicated as targets for ubiquitination by MAGE-F1-NSE1 are shown in green. e, Close-up view of the CIAO2B-CIAO2B dimer interface. Potential Fe-S cluster-coordinating residues H85, C86, and H119 are shown as sticks. Uncropped blot images for panel b are available as source data.
Extended Data Fig. 3
MMS19 sequence alignment used to map evolutionary conservation in Fig. 2b.
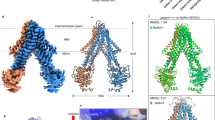
Extended Data Fig. 4 Cryo-EM reconstruction of a CTC-primase complex.
a, Size exclusion chromatography profile of assembled CTC-primase complex. b, Coomassie-stained SDS-PAGE of CTC-primase complex. c, Negative stain 2D class averages of the CTC-primase complex. d, Cryo-EM micrograph of CTC-primase sample. e, Representative 2D class averages of the CTC-primase data set. Scale bar = 100 Å. f, Overview of data processing and classification scheme. g, Angular distribution of particle orientations in the reconstruction. h, FSC plot for half-maps of the reconstruction, 0.143 FSC criterion is indicated.
Extended Data Fig. 5 Cryo-EM reconstruction of a CTC-DNA2 complex.
a, Size exclusion chromatography profile of assembled CTC-DNA2 complex. b, Coomassie-stained SDS-PAGE of CTC-DNA2 complex. c, Negative stain 2D class averages of the CTC-DNA2 complex. d, Cryo-EM micrograph of CTC-DNA2 sample. e, Representative 2D class averages of the CTC-DNA2 data set. Scale bar = 100 Å. f, Overview of data processing and classification scheme. g, Angular distribution of particle orientations in the reconstruction. h, FSC plot for half-maps of the reconstruction, 0.143 FSC criterion is indicated.
Extended Data Fig. 6 Pull-down assays in mammalian cells pinpoint blade 3 in CIAO1 as the main interaction site for recruitment of client proteins.
a, Co-expression and co-IP of Flag-MMS19 with myc-CIAO2B, wild-type or mutant myc-CIAO1, and either myc-PriL/S or myc-DNA2 in HEK293 cells. b, Location of mutated amino acid residues around blade 3 of CIAO1. The inset shows the location of the conserved and charged patches in blade 3 of CIAO1. Uncropped blot images for panel a are available as source data.
Extended Data Fig. 7 Compositional dynamics of the CTC and its potential role in Fe-S cluster transfer.
a, Compositional dynamics of the CTC might not only be required for maturation of different client proteins (maturation of certain cytosolic proteins by the core complex, and of DNA metabolism proteins by the complete CTC), but also play a role in Fe-S cluster coordination and transfer through CTC dimerization. b, Model of Fe-S cluster transfer and client protein binding by the CTC. See Supplementary Note 1.
Supplementary information
Supplementary Information
Supplementary Note 1, Fig. 1 and Tables 1–3.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 3
Source data for ITC experiments.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Kassube, S.A., Thomä, N.H. Structural insights into Fe–S protein biogenesis by the CIA targeting complex. Nat Struct Mol Biol 27, 735–742 (2020). https://doi.org/10.1038/s41594-020-0454-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0454-0