Abstract
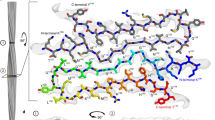
Prion diseases are caused by the misfolding of prion protein (PrP). Misfolded PrP forms protease-resistant aggregates in vivo (PrPSc) that are able to template the conversion of the native form of the protein (PrPC), a property shared by in vitro–produced PrP fibrils. Here we produced amyloid fibrils in vitro from recombinant, full-length human PrPC (residues 23–231) and determined their structure using cryo-EM, building a model for the fibril core comprising residues 170−229. The PrP fibril consists of two protofibrils intertwined in a left-handed helix. Lys194 and Glu196 from opposing subunits form salt bridges, creating a hydrophilic cavity at the interface of the two protofibrils. By comparison with the structure of PrPC, we propose that two α-helices in the C-terminal domain of PrPC are converted into β-strands stabilized by a disulfide bond in the PrP fibril. Our data suggest that different PrP mutations may play distinct roles in modulating the conformational conversion.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Prusiner, S. B. Prions. Proc. Natl Acad. Sci. USA 95, 13363–13383 (1998).
Prusiner, S. B. Molecular biology and pathogenesis of prion diseases. Trends Biochem. Sci. 21, 482–487 (1996).
Scheckel, C. & Aguzzi, A. Prions, prionoids and protein misfolding disorders. Nat. Rev. Genet. 19, 405–418 (2018).
Watts, J. C., Bourkas, M. E. C. & Arshad, H. The function of the cellular prion protein in health and disease. Acta Neuropathol. 135, 159–178 (2018).
Kim, M.-O., Takada, L. T., Wong, K., Forner, S. A. & Geschwind, M. D. Genetic PrP prion diseases. Cold Spring Harb. Perspect. Biol. 10, a033134 (2018).
Pan, K. M. et al. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl Acad. Sci. USA 90, 10962–10966 (1993).
Soto, C. Prion hypothesis: the end of the controversy? Trends Biochem. Sci. 36, 151–158 (2011).
Prusiner, S. B. A unifying role for prions in neurodegenerative diseases. Science 336, 1511–1513 (2012).
Rossetti, G., Cong, X., Caliandro, R., Legname, G. & Carloni, P. Common structural traits across pathogenic mutants of the human prion protein and their implications for familial prion diseases. J. Mol. Biol. 411, 700–712 (2011).
Diaz-Espinoza, R. & Soto, C. High-resolution structure of infectious prion protein: the final frontier. Nat. Struct. Mol. Biol. 19, 370–377 (2012).
Soto, C., Estrada, L. & Castilla, J. Amyloids, prions and the inherent infectious nature of misfolded protein aggregates. Trends Biochem. Sci. 31, 150–155 (2006).
Zahn, R. et al. NMR solution structure of the human prion protein. Proc. Natl Acad. Sci. USA 97, 145–150 (2000).
Spagnolli, G. et al. Full atomistic model of prion structure and conversion. PLoS Pathog. 15, e1007864 (2019).
Wille, H. et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc. Natl Acad. Sci. USA 106, 16990–16995 (2009).
Smirnovas, V. et al. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 18, 504–506 (2011).
Gallagher-Jones, M. et al. Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp. Nat. Struct. Mol. Biol. 25, 131–134 (2018).
Govaerts, C., Wille, H., Prusiner, S. B. & Cohen, F. E. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl Acad. Sci. USA 101, 8342–8347 (2004).
Vázquez-Fernández, E. et al. The structural architecture of an infectious mammalian prion using electron cryomicroscopy. PLoS Pathog. 12, e1005835 (2016).
Lu, X., Wintrode, P. L. & Surewicz, W. K. β-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc. Natl Acad. Sci. USA 104, 1510–1515 (2007).
Cobb, N. J., Sönnichsen, F. D., Mchaourab, H. & Surewicz, W. K. Molecular architecture of human prion protein amyloid: a parallel, in-register β-structure. Proc. Natl Acad. Sci. USA 104, 18946–18951 (2007).
Terry, C. et al. Structural features distinguishing infectious ex vivo mammalian prions from non-infectious fibrillar assemblies generated in vitro. Sci. Rep. 9, 376 (2019).
Bocharova, O. V., Breydo, L., Parfenov, A. S., Salnikov, V. V. & Baskakov, I. V. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrPSc. J. Mol. Biol. 346, 645–659 (2005).
Tattum, M. H. et al. Elongated oligomers assemble into mammalian PrP amyloid fibrils. J. Mol. Biol. 357, 975–985 (2006).
Zhou, Z. et al. Fibril formation of the rabbit/human/bovine prion proteins. Biophys. J. 101, 1483–1492 (2011).
Pan, K., Yi, C. W., Chen, J. & Liang, Y. Zinc significantly changes the aggregation pathway and the conformation of aggregates of human prion protein. Biochim. Biophys. Acta 1854, 907–918 (2015).
Legname, G. et al. Synthetic mammalian prions. Science 305, 673–676 (2004).
Colby, D. W. et al. Protease-sensitive synthetic prions. PLoS Pathog. 6, e1000736 (2010).
Colby, D. W. et al. Design and construction of diverse mammalian prion strains. Proc. Natl Acad. Sci. USA 106, 20417–20422 (2009).
Yang, F. Jr., Zhang, M., Zhou, B. R., Chen, J. & Liang, Y. Oleic acid inhibits amyloid formation of the intermediate of α-lactalbumin at moderately acidic pH. J. Mol. Biol. 362, 821–834 (2006).
Terry, C. et al. Ex vivo mammalian prions are formed of paired double helical prion protein fibrils. Open Biol. 6, 160035 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Falcon, B. et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 (2019).
Bocharova, O. V., Breydo, L., Salnikov, V. V. & Baskakov, I. V. Copper(II) inhibits in vitro conversion of prion protein into amyloid fibrils. Biochemistry 44, 6776–6787 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Terwilliger, T. C., Sobolev, O. V., Afonine, P. V. & Adams, P. D. Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr. D Struct. Biol. 74, 545–559 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
Y.L. and C.L. acknowledge funding from the National Natural Science Foundation of China (no. 31770833) and the Major State Basic Research Development Program (no. 2016YFA0501902). Y.L. also acknowledges financial support from the National Natural Science Foundation of China (nos. 31570779 and 31370774), the National Key Basic Research Foundation of China (no. 2013CB910702) and the Fundamental Research Fund for the Central Universities of China (no. 2015204020201). C.L. was also supported by the National Natural Science Foundation of China (no. 91853113), the Science and Technology Commission of Shanghai Municipality (no. 18JC1420500) and Shanghai Municipal Science and Technology Major Project (no. 2019SHZDZX02). P.Y. acknowledges financial support from the Major State Basic Research Development Program (no. 2018YFA0507700) and the National Natural Science Foundation of China (no. 31722017). Cryo-EM data were collected at the Center of Cryo Electron Microscopy, Zhejiang University, China. We thank G.-F. Xiao (Wuhan Institute of Virology, Chinese Academy of Sciences) for the kind gift of the human PrPC plasmid; S. Chang (Center of Cryo Electron Microscopy, Zhejiang University) and X. Zhang (Zhejiang University School of Medicine) for their technical assistance with Cryo-EM; and Y. Wang (Institute of Biophysics, Chinese Academy of Sciences) for her helpful suggestions.
Author information
Authors and Affiliations
Contributions
P.Y., C.L. and Y.L. supervised the project. L.-Q.W., P.Y., C.L. and Y.L. designed the experiments. L.-Q.W., H.-Y.Y., J.T., X.-N.L. and J.C. purified the PrPC and PrP fibrils. L.-Q.W. and C.-W.Y. performed Congo red binding and proteinase K digestion assays of PrP fibrils. L.-Q.W., K.Z., H.-Y.Y., Q.W., Z.G., D.Z., Y.S. and D.L. collected, processed and/or analyzed cryo-EM data. L.-Q.W., K.Z., C.L. and Y.L. wrote the manuscript. All authors proofread and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
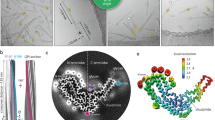
Extended Data Fig. 1 PrP fibrils bind to Congo red and are proteinase K resistant.
a, Amyloid fibrils of full-length human PrP analyzed by Congo red binding assays (a) or concentration-dependent proteinase K digestion assays (b)24,25. a, The difference spectra (Curve 4, blue) with the maximum absorbance at 550 nm were obtained by subtracting the absorbance spectra of PrP fibrils alone (Curve 3, black) and Congo red alone (Curve 1, red) with the maximum absorbance at 495 nm from those of PrP fibrils + Congo red (Curve 2, green). Congo red binding assays were carried out at 37 °C. b, Protease-resistant core fragment of 15−16-kDa is highlighted using a black arrow. Samples were treated with proteinase K for 1 h at 37 °C at protease:PrP molar ratios of 1:100 (lane 3) and 1:50 (lane 4). The control with no protease was loaded in lane 1. Molecular weight markers were loaded on lane 2: restriction endonuclease Bsp98 I (25.0 kDa), β-lactoglobulin (18.4 kDa), and lysozyme (14.4 kDa). Protein fragments were separated by SDS-PAGE and detected by silver staining. These experiments were repeated three times with different batches of fibrils and similar results. Data behind graph and uncropped images are available as source data.
Extended Data Fig. 2 Cryo-EM images of PrP fibrils.
a, Cryo-EM micrographs of amyloid fibrils of full-length human PrP showing two protofibrils intertwined into a left-handed helix. Scale bar, 50 nm. b, Reference-free 2D class averages of PrP fibrils showing two protofibrils intertwined. Scale bar, 10 nm. c, Enlarged image of (b) showing two protofibrils arranged in a staggered manner. Scale bar, 2 nm.
Extended Data Fig. 3 Global (a) and local resolution (b) estimates for the PrP fibril reconstructions.
a, Gold-standard refinement was used for estimation of the density map resolution. The global resolution of 2.70 Å was calculated using a Fourier shell correlation (FSC) curve cut-off at 0.143. b, The density map of PrP fibrils is colored according to local resolution estimated by ResMap. The three enlarged cross sections show the left top view of the density map of two protofibrils. The color key on the right shows the local structural resolution in angstroms (Å) and the colored map indicates the local resolution ranging from 2.6 to 4.5 Å.
Extended Data Fig. 4 Close-up view of the density map of PrP fibrils with the atomic model overlaid.
a, The dimer interface comprises residues Lys194, Gly195, and Glu196 from both subunits. Two salt bonds are formed between Lys194 and Glu196 from opposing subunits to create a hydrophilic cavity at the dimerization interface. b, An inner cavity in each subunit formed by Gln186, Val189, and Thr191 on one side and Val203 and Met205 on the opposing side. c, A salt bridge is formed between Glu211 and Arg228. d, PrP fibrils are stabilized by one disulfide bond between Cys179 and Cys214. A hydrogen bond is formed between His177 and Tyr218. e, Hydrophobic side chains of Val180, Ile182, Ile184, Met205, Met206, Val209, and Val210 are located in the interior of PrP fibrils to form a stable hydrophobic core. f, A U-turn between β5 and β6 containing residues 217QYERES222. Three hydrogen bonds are formed between Gln217 and the main chain of Gln223, between Ser222 and the main chain of Glu219, and between Ser222 and the main chain of Arg220.
Extended Data Fig. 5 Cryo-EM density map of human PrP fibril with the atomic model overlaid.
A cavity at the interface of two protofibrils encloses two additional densities that are not connected to PrP. An internal cavity encloses extra density that is also not connected to PrP. Three ordered solvent molecules are found in the inner cavities.
Supplementary information
Source data
Source Data Extended Data Fig. 1.
Statistical source data for Extended Data Fig. 1a.
Source Data Extended Data Fig. 1.
Full-length, unprocessed gels for Extended Data Fig. 1b.
Rights and permissions
About this article
Cite this article
Wang, LQ., Zhao, K., Yuan, HY. et al. Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat Struct Mol Biol 27, 598–602 (2020). https://doi.org/10.1038/s41594-020-0441-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0441-5
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Synthetic β-sheets mimicking fibrillar and oligomeric structures for evaluation of spectral X-ray scattering technique for biomarker quantification
Cell & Bioscience (2024)
-
Structural polymorphism of amyloid fibrils in ATTR amyloidosis revealed by cryo-electron microscopy
Nature Communications (2024)
-
Excess PrPC inhibits muscle cell differentiation via miRNA-enhanced liquid–liquid phase separation implicated in myopathy
Nature Communications (2023)
-
Prion protein amino acid sequence influences formation of authentic synthetic PrPSc
Scientific Reports (2023)