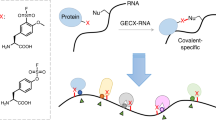
Abstract
RNA-binding sites (RBSs) can be identified by liquid chromatography and tandem mass spectrometry analyses of the protein–RNA conjugates created by crosslinking, but RBS mapping remains highly challenging due to the complexity of the formed RNA adducts. Here, we introduce RBS-ID, a method that uses hydrofluoride to fully cleave RNA into mono-nucleosides, thereby minimizing the search space to drastically enhance coverage and to reach single amino acid resolution. Moreover, the simple mono-nucleoside adducts offer a confident and quantitative measure of direct RNA–protein interaction. Using RBS-ID, we profiled ~2,000 human RBSs and probed Streptococcus pyogenes Cas9 to discover residues important for genome editing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
References
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Baltz, A. G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Leitner, A., Dorn, G. & Allain, F. H. T. Combining mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy for integrative structural biology of protein–RNA complexes. Cold Spring Harb. Perspect. Biol. 11, a032359 (2019).
Castello, A. et al. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 (2016).
He, C. et al. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416–430 (2016).
Kramer, K. et al. Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods 11, 1064–1070 (2014).
Panhale, A. et al. CAPRI enables comparison of evolutionarily conserved RNA interacting regions. Nat. Commun. 10, 2682 (2019).
Shchepachev, V. et al. Defining the RNA interactome by total RNA-associated protein purification. Mol. Syst. Biol. 15, e8689 (2019).
Jeong, K., Kim, S. & Bandeira, N. False discovery rates in spectral identification. BMC Bioinformatics 13(Suppl. 16), S2 (2012).
Bogdanow, B., Zauber, H. & Selbach, M. Systematic errors in peptide and protein identification and quantification by modified peptides. Mol. Cell Proteom. 15, 2791–2801 (2016).
Trendel, J. et al. The human RNA-binding proteome and its dynamics during translational arrest. Cell 176, 391–403 (2019).
Crean, C., Uvaydov, Y., Geacintov, N. E. & Shafirovich, V. Oxidation of single-stranded oligonucleotides by carbonate radical anions: generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 36, 742–755 (2008).
Woo, E. M., Fenyo, D., Kwok, B. H., Funabiki, H. & Chait, B. T. Efficient identification of phosphorylation by mass spectrometric phosphopeptide fingerprinting. Anal. Chem. 80, 2419–2425 (2008).
Na, S., Bandeira, N. & Paek, E. Fast multi-blind modification search through tandem mass spectrometry. Mol. Cell Proteom. 11, 010199 (2012).
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Kim, S. & Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277 (2014).
Edwards, N. J. PepArML: a meta-search peptide identification platform for tandem mass spectra. Curr. Protoc. Bioinformatics 44, 13.23.1–13.23.23 (2013).
Chalkley, R. J. & Clauser, K. R. Modification site localization scoring: strategies and performance. Mol. Cell Proteom. 11, 3–14 (2012).
Chang, C. et al. PANDA: a comprehensive and flexible tool for quantitative proteomics data analysis. Bioinformatics 35, 898–900 (2019).
UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Schafer, I. B. et al. Molecular basis for poly(A) RNP architecture and recognition by the Pan2-Pan3 deadenylase. Cell 177, 1619–1631 (2019).
Kuhn, U. & Pieler, T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein–protein interaction. J. Mol. Biol. 256, 20–30 (1996).
Hawley, B. R., Lu, W. T., Wilczynska, A. & Bushell, M. The emerging role of RNAs in DNA damage repair. Cell Death Differ. 24, 580–587 (2017).
Shetlar, M. D., Carbone, J., Steady, E. & Hom, K. Photochemical addition of amino acids and peptides to polyuridylic acid. Photochem. Photobiol. 39, 141–144 (1984).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Yoon, J. H. et al. Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs. Nucleic Acids Res. 42, 1196–1208 (2014).
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J. A. A Cas9–guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Leonetti, M. D., Sekine, S., Kamiyama, D., Weissman, J. S. & Huang, B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl Acad. Sci. USA 113, E3501–3508 (2016).
He, L., Diedrich, J., Chu, Y. Y. & Yates, J. R. III. Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal. Chem. 87, 11361–11367 (2015).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Vizcaino, J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013).
Acknowledgements
We thank S. Shin, K. Baeg and S. Lee for insightful comments and discussion. We are also grateful to J. Kim, J. Yang, D. Choi and E. Kim for technical help, and all members of our laboratories for helpful discussion. We thank J. S. Kim (Seoul National University), Pacific Northwest National Laboratory and the OMICS.PNL.GOV for providing valuable plasmid and software. This work was supported by IBS-R008-D1 of the Institute for Basic Science from the Ministry of Science and ICT of Korea (J.W.B., S.-C.K., Y.N., V.N.K. and J.-S.K.) and BK21 Research Fellowships (J.W.B.) from the Ministry of Education, Science and Technology of Korea.
Author information
Authors and Affiliations
Contributions
J.W.B., V.N.K. and J.-S.K. conceived the project and designed the experiments. J.W.B. developed the protocol and performed all biochemical experiments with the support of S.C.K. and Y.N. J.W.B. generated and analyzed all LC-MS/MS datasets. J.W.B., V.N.K. and J.-S.K. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available. Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 HF treatment on RNA and peptides.
a, Schematic illustration of HF treatment on RNA (left) and the resulting products (right). HF-mediated cleavage sites are highlighted. b, UV-absorbance chromatogram of 20-mer RNA (AUGCAUGCAUGCAUGCAUGC) digested with HF (black solid and dashed lines from duplicate experiments), merged with those of undigested RNA (gray) and reference chemicals (colored, solid). Peaks of reference chemicals were re-sized for better visualization. The black solid line is the source data for Fig. 1b. c, UV-absorbance chromatogram of HeLaT total RNA digested with HF (black, solid or dashed lines from duplicate experiments), merged with those of reference chemicals (colored, solid). Peaks of reference chemicals were re-sized for better visualization. d, Proportion of semi-tryptic PSM identified from HeLaT digest peptides upon HF treatment, compared to negative control treated with H2O. Two-sided unpaired Student’s T-test was performed between n = 3 biologically independent samples (H2O vs. HF), assuming equal variance. The mean values were depicted with error bars that indicate standard deviation between replicates. P-value, rounded up to the fourth decimal point: 0.0258. e, Proportion of identified PSM (below PSM-level FDR = 0.01) upon HF treatment, compared to H2O treatment. Two-sided unpaired Student’s T-test was performed as described above. The mean values were depicted with error bars that indicate standard deviation between replicates. P-value, rounded up to the fourth decimal point: 0.1359.
Extended Data Fig. 2 Open search on total RNA-RBS and mRNA-RBS using MODa and MSFragger.
a, MODa14 search for modified mass on mRNA-RBS. The y-axis indicates mean spectral counts from duplicate experiments. b-c, MSFragger15 search for modified mass on total RNA-RBS (b) and mRNA-RBS (c). d, Mean percentage of Uracil modification over Uridine modification, a highly likely in-source fragmentation product, between replicate experiments, calculated from MSFragger search results as (#PSM with Uracil adduct: 112 & 55 to account for Cys)/(#PSM with Uracil or Uridine adduct: 244, 112 & 187, 55 to account for Cys). The error bars indicate standard deviation. Free Cys was carbamidomethylated, so the corresponding adduct mass was used as a fixed modification. Owing to mutually exclusive U-crosslinking and carbamidomethylation on Cys, the observed conjugate mass of uridine on Cys (187) was smaller than that of other amino acids (244) by the difference of mass of carbamidomethyl group (57). Thus, modification mass on Cys was corrected by adding the mass of carbamidomethyl group. The percentages were rounded up to the second decimal point. e, Comparison of closed search results on total RNA and mRNA RBS-ID experiments, allowing up to one or two modifications per peptide. Modification-specific peptide-level FDR was set to 0.01. PSM counts for peptides with two modifications were depicted.
Extended Data Fig. 3 Comparison between RBS-ID and previous RBS- or RBD-profiling studies.
a, Comparison of the position of RBS in peptides shared between RBS-ID and RNPxl Venn diagram (left) shows the overlap between the peptides with uniquely localized RBS in RBS-ID (1,972) and those in human proteins of RNPxl (29)6. Please note that most peptides from RNPxl are also detected by RBS-ID, demonstrating the comprehensiveness of our method. The Pie chart (right) displays that among 25 common peptides, 24 peptides show consistent localization of RBS between the datasets, indicating the accuracy of the methods. b, Relative position of the peptides identified as ‘RBDpep’4. X-axis shows the position of the terminus of peptides relative to RBSs identified by RBS-ID (n = 1,478). c, Relative position of the peptides identified as ‘XL-peptide’7. X-axis shows the position of the terminus of peptides relative to RBSs identified by RBS-ID (n = 869).
Extended Data Fig. 4 Reproducibility of MS1 intensity-based label-free quantification.
a-b, MS1 intensity-based label-free quantification27 of RBS-containing peptides co-identified in Total RNA-RBS (a) or mRNA-RBS (b) replicate experiments. Pearson’s correlation coefficient and Spearman’s correlation coefficient were each calculated and rounded up to the second decimal point.
Extended Data Fig. 5 RBS-identified protein groups and regions.
a, Top 5 GO terms associated with proteins that are not annotated as RBPs (MF: molecular function, BP: biological process, CC: Cellular component, 5 each)20,21. b-c, Top 5 GO terms associated with proteins whose RBSs are identified exclusively in total RNA enrichment but not in poly(A) + RNA enrichment (b) or in poly(A) + RNA enrichment but not in total RNA enrichment (c) (MF, BP, CC, 5 each).
Extended Data Fig. 6 Examples of RBS-identified proteins.
a, Example of RBSs identified in distant primary sequence positions (PABPC1). Sequence homology of amino acids at -5 to +5 positions from Y194, Y297, and Y364 compared to that of Y222, F325, and Y393 in yeast Pab1 are described, respectively. Identical amino acids in the same positions are bold-faced. b, Partial structure of yeast Pab1 bound to poly(A) RNA (PDB 6R5K22). Y222, F325, and Y393 are indicated. c-h, Examples of RBSs identified in regions that are not annotated as RBDs. RBSs identified in NSUN5 (c), RTCA (d), APOBEC3C (e), TRIM25 (f), SERBP1 (g), and HNRNPA1 (h) are depicted. RBSs with high spectral counts are indicated.
Extended Data Fig. 7 Purified spCas9 protein and template DNA for sgRNA synthesis.
a, Purified His6-HA-NLS-TEV-spCas9 on SDS-PAGE gel stained with Coomassie G25. b, Template DNA prepared for T7 in vitro transcription of anti-CBX1 sgRNA.
Extended Data Fig. 8 Positions of RBS in crystal structure of spCas9 in complex with sgRNA and target DNA.
Positions of RBSs in the crystal structure of spCas9 in complex with sgRNA and target DNA (PDB 4UN329). Individual atoms of RBSs were drawn as dark blue spheres.
Extended Data Fig. 9 Impact of RBS mutagenesis on spCas9 gene editing activity.
a, EtBr-agarose gel image of cut/uncut fragments after T7E1 assay30, 48 hours after transfection of negative control (mock), non-target-wildtype (NC), wildtype (WT), Y450A, R919A spCas9. b, EtBr-agarose gel image of input PCR amplicons. c, Histograms of GFP fluorescence of msfGFP-expressing clonal stable 293 cells 96 hours after transfection with non-target-wildtype (NC), wildtype (WT), Y450A, R919A spCas9. d, Cell counts per each flow cytometry run. Uncropped images for panels a-b are available as source data.
Extended Data Fig. 10 Verification of comparable expression of spCas9.
a, Verification of comparable expression of spCas9 proteins. Western blot of msfGFP-expressing HEK293E clonal stable cell 48 hours after transfection of negative control (mock), non-target-wildtype (NC), wildtype (WT), Y450A, or R919A spCas9. b, Quantification of spCas9 protein expression level. Two-sided unpaired Student’s T-test was performed with n = 3 biologically independent samples (WT vs. Y450A; WT vs. R919A), assuming equal variance. The mean values were depicted with error bars that indicate standard deviation between replicates. P-value, rounded up to the fourth decimal point: 0.3261 (WT vs. Y450A), 0.2257 (WT vs. R919A). Uncropped image for panel a is available as source data.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3.
Supplementary Table 1
Human RBS open search results.
Supplementary Table 2
Human RBS closed search results.
Supplementary Table 3
spCas9 RBS closed search results.
Source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 9
Unprocessed gels
Source Data Extended Data Fig. 10
Unprocessed western blots
Rights and permissions
About this article
Cite this article
Bae, J.W., Kwon, S.C., Na, Y. et al. Chemical RNA digestion enables robust RNA-binding site mapping at single amino acid resolution. Nat Struct Mol Biol 27, 678–682 (2020). https://doi.org/10.1038/s41594-020-0436-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0436-2
This article is cited by
-
Structure-based prediction and characterization of photo-crosslinking in native protein–RNA complexes
Nature Communications (2024)
-
RNA-binding proteins in cardiovascular biology and disease: the beat goes on
Nature Reviews Cardiology (2024)
-
RNA-binding proteins that lack canonical RNA-binding domains are rarely sequence-specific
Scientific Reports (2023)
-
Nucleotide-amino acid π-stacking interactions initiate photo cross-linking in RNA-protein complexes
Nature Communications (2022)
-
The RNA-bound proteome of MRSA reveals post-transcriptional roles for helix-turn-helix DNA-binding and Rossmann-fold proteins
Nature Communications (2022)