Abstract
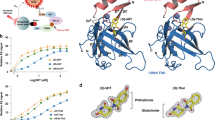
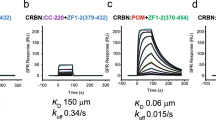
Thalidomide-dependent degradation of the embryonic transcription factor SALL4 by the CRL4CRBN E3 ubiquitin ligase is a plausible major driver of thalidomide teratogenicity. The structure of the second zinc finger of SALL4 in complex with pomalidomide, cereblon and DDB1 reveals the molecular details of recruitment. Sequence differences and a shifted binding position relative to Ikaros offer a path to the rational design of cereblon-binding drugs with reduced teratogenic risk.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Vargesson, N. The teratogenic effects of thalidomide on limbs. J. Hand Surg. Eur. Vol. 44, 88–95 (2019).
Chamberlain, P. P. & Hamann, L. G. Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 15, 937–944 (2019).
Matyskiela, M. E. et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535, 252–257 (2016).
Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010).
Angers, S. et al. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 443, 590–593 (2006).
Chamberlain, P. P. et al. Structure of the human Cereblon–DDB1–lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat. Struct. Mol. Biol. 21, 803–809 (2014).
Fischer, E. S. et al. Structure of the DDB1–CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
An, J. et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nat. Commun. 8, 15398 (2017).
Donovan, K. A., et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife 7, e38430 (2018).
Kronke, J. et al. Lenalidomide induces ubiquitination and degradation of CK1ɑ in del(5q) MDS. Nature 523, 183–188 (2015).
Kronke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014).
Petzold, G., Fischer, E. S. & Thoma, N. H. Structural basis of lenalidomide-induced CK1ɑ degradation by the CRL4(CRBN) ubiquitin ligase. Nature 532, 127–130 (2016).
Sievers, Q.L., et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362, eaat0572 (2018).
Gandhi, A. K. et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br. J. Haematol. 164, 811–821 (2014).
Matyskiela, M. E. et al. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat. Chem. Biol. 14, 981–987 (2018).
Kohlhase, J. & Holmes, L. B. Mutations in SALL4 in malformed father and daughter postulated previously due to reflect mutagenesis by thalidomide. Birth Defects Res. A Clin. Mol. Teratol. 70, 550–551 (2004).
Kohlhase, J. et al. Mutations at the SALL4 locus on chromosome 20 result in a range of clinically overlapping phenotypes, including Okihiro syndrome, Holt–Oram syndrome, acro-renal-ocular syndrome, and patients previously reported to represent thalidomide embryopathy. J. Med. Genet. 40, 473–478 (2003).
Borozdin, W. et al. SALL4 deletions are a common cause of Okihiro and acro-renal-ocular syndromes and confirm haploinsufficiency as the pathogenic mechanism. J. Med. Genet. 41, e113 (2004).
Asatsuma-Okumura, T. et al. p63 is a cereblon substrate involved in thalidomide teratogenicity. Nat. Chem. Biol. 15, 1077–1084 (2019).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. et al. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Harder, E. et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296 (2016).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
Thanks to M. Riley for baculovirus expression support and J. Pack, M. Ellis and L. LeBrun for thoughtful comments on the manuscript. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
T.C., E.T., B.P., A.C., M.E.M. and P.P.C. performed crystallographic studies and in vitro assays. X.Z. and G.L. performed immunoprecipitation experiments. C.M. and J.M. performed molecular dynamics simulations. M.R., L.G.H. and P.P.C. planned the research.
Corresponding authors
Ethics declarations
Competing interests
The authors are, or have been, employees or contractors of Celgene and/or Bristol-Myers Squibb.
Additional information
Peer review information Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sample electron density from the cereblon-DDB1-SALL4 ZF2-pomalidomide crystal structure.
Fo-Fc electron density map calculated in the absence of pomalidomide and contoured at 3σ following a round of refinement. Protein atoms are not shown in the figure.
Extended Data Fig. 2 SALL4 binding defects upon cereblon mutation mapped onto the structure of cereblon.
The surface of cereblon is colored by the effect of alanine mutations at that position on SALL4 ternary complex formation (blue, untested; green, no effect; red, detrimental effect; magenta, increased complex formation).
Extended Data Fig. 3 Structural model of SALL4 recruitment by the CRL4CRBN complex.
Structural model of SALL4 recruitment by pomalidomide to the cereblon-CRL4 E3 ligase complex, positioning SALL4 nearby the site of E2 recruitment on RBX11.
Supplementary information
Supplementary Information
Supplementary Table 1.
Source data
Source Data Fig. 2
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Matyskiela, M.E., Clayton, T., Zheng, X. et al. Crystal structure of the SALL4–pomalidomide–cereblon–DDB1 complex. Nat Struct Mol Biol 27, 319–322 (2020). https://doi.org/10.1038/s41594-020-0405-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0405-9
This article is cited by
-
SALL4 is a CRL3REN/KCTD11 substrate that drives Sonic Hedgehog-dependent medulloblastoma
Cell Death & Differentiation (2024)
-
DNA-encoded library-enabled discovery of proximity-inducing small molecules
Nature Chemical Biology (2024)
-
Novel, thalidomide-like, non-cereblon binding drug tetrafluorobornylphthalimide mitigates inflammation and brain injury
Journal of Biomedical Science (2023)
-
Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon
Scientific Reports (2023)
-
Defining molecular glues with a dual-nanobody cannabidiol sensor
Nature Communications (2022)