Abstract
The prevalent model for cataract formation in the eye lens posits that damaged crystallin proteins form light-scattering aggregates. The α-crystallins are thought to counteract this process as chaperones by sequestering misfolded crystallin proteins. In this scenario, chaperone pool depletion would result in lens opacification. Here we analyze lenses from different mouse strains that develop early-onset cataract due to point mutations in α-, β-, or γ-crystallin proteins. We find that these mutant crystallins are unstable in vitro; in the lens, their levels are substantially reduced, and they do not accumulate in the water-insoluble fraction. Instead, all the other crystallin proteins, including the α-crystallins, are found to precipitate. The changes in protein composition and spatial organization of the crystallins observed in the mutant lenses suggest that the imbalance in the lenticular proteome and altered crystallin interactions are the bases for cataract formation, rather than the aggregation propensity of the mutant crystallins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the conclusions are available with the article. Raw data, including polyacrylamide gels, are available upon request to the authors.
References
Petrash, J. M. Aging and age-related diseases of the ocular lens and vitreous body. Invest. Ophthalmol. Vis. Sci. 54, 54–59 (2013).
Ainsbury, E. A. et al. Ionizing radiation induced cataracts: recent biological and mechanistic developments and perspectives for future research. Mutat. Res. 770, 238–261 (2016).
Duncan, M. K., Cvekl, A., Kantorow, M. & Piatigorsky, J. in Development of the Ocular Lens (eds Lovicu, F. J. & Robinson, M. L.) Ch. 5 (Cambridge University Press, 2004).
Löfgren, S. Solar ultraviolet radiation cataract. Exp. Eye Res. 156, 112–116 (2017).
Graw, J. The genetic and molecular basis of congenital eye defects. Nat. Rev. Genet. 4, 876–888 (2003).
Michael, R. & Bron, A. J. The ageing lens and cataract: a model of normal and pathological ageing. Philos. Trans. R. Soc. Lond. B Bio. Sci. 366, 1278–1292 (2011).
Churchill, A. & Graw, J. Clinical and experimental advances in congenital and paediatric cataracts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 1234–1249 (2011).
Graw, J. Genetics of crystallins: cataract and beyond. Exp. Eye Res. 88, 173–189 (2009).
Truscott, R. J. Age-related nuclear cataract-oxidation is the key. Exp. Eye Res. 80, 709–725 (2005).
Zhang, Z., Smith, D. L. & Smith, J. B. Human β-crystallins modified by backbone cleavage, deamidation and oxidation are prone to associate. Exp. Eye Res. 77, 259–272 (2003).
Takemoto, L. & Sorensen, C. M. Protein–protein interactions and lens transparency. Exp. Eye Res. 87, 496–501 (2008).
Schaefer, H. et al. Study of posttranslational modifications in lenticular αA-crystallin of mice using proteomic analysis techniques. Biochim. Biophys. Acta 1764, 1948–1962 (2006).
Carver, J. A., Ecroyd, H., Truscott, R. J. W., Thorn, D. C. & Holt, C. Proteostasis and the regulation of intra- and extracellular protein aggregation by ATP-independent molecular chaperones: lens α-crystallins and milk caseins. Acc. Chem. Res. 51, 745–752 (2018).
Sandilands, A. et al. Altered aggregation properties of mutant γ-crystallins cause inherited cataract. EMBO J. 21, 6005–6014 (2002).
Lee, S. et al. A single destabilizing mutation (F9S) promotes concerted unfolding of an entire globular domain in γS-crystallin. J. Mol. Biol. 399, 320–330 (2010).
World Report on Vision (WHO, 2019).
Takemoto, L. & Boyle, D. The possible role of α-crystallins in human senile cataractogenesis. Int. J. Biol. Macromol. 22, 331–337 (1998).
Delaye, M. & Tardieu, A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature 302, 415–417 (1983).
Tardieu, A. Eye lens proteins and transparency: from light transmission theory to solution X-ray structural analysis. Annu. Rev. Biophys. Biophys. Chem. 17, 47–70 (1988).
Tardieu, A. α-Crystallin quaternary structure and interactive properties control eye lens transparency. Int. J. Biol. Macromol. 22, 211–217 (1998).
Jaenicke, R. & Slingsby, C. Lens crystallins and their microbial homologs: structure, stability, and function. Crit. Rev. Biochem. Mol. Biol. 36, 435–499 (2001).
Clark, A. R., Lubsen, N. H. & Slingsby, C. sHSP in the eye lens: crystallin mutations, cataract and proteostasis. Int. J. Biochem. Cell Biol. 44, 1687–1697 (2012).
Moreau, K. L. & King, J. A. Protein misfolding and aggregation in cataract disease and prospects for prevention. Trends Mol. Med. 18, 273–282 (2012).
Day, T. H. & Clayton, R. M. Multiple changes in lens protein composition associated with CatFr gene in the mouse. Genet. Res. 19, 241–249 (1972).
Berthoud, V. M. et al. Connexin50D47A decreases levels of fiber cell connexins and impairs lens fiber cell differentiation. Invest. Ophthalmol. Vis. Sci. 54, 7614–7622 (2013).
Posner, M., McDonald, M. S., Murray, K. L. & Kiss, A. J. Why does the zebrafish cloche mutant develop lens cataract? PLoS ONE 14, e0211399 (2019).
Graw, J. et al. Characterization of a new, dominant V124E mutation in the mouse αA-crystallin-encoding gene. Invest. Ophthalmol. Vis. Sci. 42, 2909–2915 (2001).
Puk, O., Ahmad, N., Wagner, S., Hrabé de Angelis, M. & Graw, J. First mutation in the βA2-crystallin encoding gene is associated with small lenses and age-related cataracts. Invest. Ophthalmol. Vis. Sci. 52, 2571–2576 (2011).
Graw, J. et al. V76D mutation in a conserved γD-crystallin region leads to dominant cataracts in mice. Mamm. Genome 13, 452–455 (2002).
Santhoshkumar, P., Xie, L., Raju, M., Reneker, L. & Sharma, K. K. Lens crystallin modifications and cataract in transgenic mice overexpressing acylpeptide hydrolase. J. Biol. Chem. 289, 9039–9052 (2014).
Lyon, Y. A., Sabbah, G. M. & Julian, R. R. Differences in α-crystallin isomerization reveal the activity of protein isoaspartyl methyltransferase (PIMT) in the nucleus and cortex of human lenses. Exp. Eye Res. 171, 131–141 (2018).
Lund, A. L., Smith, J. B. & Smith, D. L. Modifications of the water-insoluble human lens α-crystallins. Exp. Eye Res. 63, 661–672 (1996).
Lyon, Y. A. et al. Structural and functional consequences of age-related isomerization in α-crystallins. J. Biol. Chem. 294, 7546–7555 (2019).
Horwitz, J., Bova, M. P., Ding, L.-L., Haley, D. A. & Stewart, P. L. Lens α-crystallin: function and structure. Eye (Lond.) 13, 403–408 (1999).
Ueda, Y., Duncan, M. K. & David, L. L. Lens proteomics: the accumulation of crystallin modifications in the mouse lens with age. Invest. Ophthalmol. Vis. Sci. 43, 205–215 (2002).
Fan, J. et al. A role for γS-crystallin in the organization of actin and fiber cell maturation in the mouse lens. FEBS J. 279, 2892–2904 (2012).
Lampi, K. J., Shih, M., Ueda, Y., Shearer, T. R. & David, L. L. Lens proteomics: analysis of rat crystallin sequences and two-dimensional electrophoresis map. Invest. Ophthalmol. Vis. Sci. 43, 216–224 (2002).
Jungblut, P. R. et al. Identification of mouse crystallins in 2D protein patterns by sequencing and mass spectrometry. Application to cataract mutants. FEBS Lett. 435, 131–137 (1998).
Puk, O., Hrabé de Angelis, M. & Graw, J. Lens density tracking in mice by Scheimpflug imaging. Mamm. Genome 24, 295–302 (2013).
Leveille, P. J., Weindruch, R., Walford, R. L., Bok, D. & Horwitz, J. Dietary restriction retards age-related loss of gamma crystallins in the mouse lens. Science 224, 1247–1249 (1984).
Ryazantsev, S. N., Poliansky, N. B., Chebotareva, N. A. & Muranov, K. O. 3D structure of the native α-crystallin from bovine eye lens. Int. J. Biol. Macromol. 117, 1289–1298 (2018).
Lam, D. et al. Cataract. Nat. Rev. Dis. Primers 1, 15014 (2015).
Cai, J., Townsend, J. P., Dodson, T. C., Heiney, P. A. & Sweeney, A. M. Eye patches: protein assembly of index-gradient squid lenses. Science 357, 564–569 (2017).
Mirarefi, A. Y. et al. Small-angle X-ray scattering studies of the intact eye lens: effect of crystallin composition and concentration on microstructure. Biochim. Biophys. Acta 1800, 556–564 (2010).
Moreau, K. L. & King, J. Hydrophobic core mutations associated with cataract development in mice destabilize human γD-crystallin. J. Biol. Chem. 284, 33285–33295 (2009).
Graw, J. et al. Aey2, a new mutation in the βB2-crystallin-encoding gene of the mouse. Invest. Ophthalmol. Vis. Sci. 42, 1574–1580 (2001).
Kuck, J. F., Kuwabara, T. & Kuck, K. D. The Emory mouse cataract: an animal model for human senile cataract. Curr. Eye Res. 1, 643–649 (1981).
Alperstein, A. M., Ostrander, J. S., Zhang, T. O. & Zanni, M. T. Amyloid found in human cataracts with two-dimensional infrared spectroscopy. Proc. Natl Acad. Sci. USA 116, 6602–6607 (2019).
Fu, L. & Liang, J. J. Detection of protein-protein interactions among lens crystallins in a mammalian two-hybrid system assay. J. Biol. Chem. 277, 4255–4260 (2002).
Takemoto, L., Ponce, A. & Sorensen, C. M. Age-dependent association of γ-crystallins with aged α-crystallins from old bovine lens. Mol. Vis. 14, 970–974 (2008).
Fu, L. & Liang, J. J. Alteration of protein–protein interactions of congenital cataract crystallin mutants. Invest. Ophthalmol. Vis. Sci. 44, 1155–1159 (2003).
Makley, L. N. et al. Pharmacological chaperone for α-crystallin partially restores transparency in cataract models. Science 350, 674–677 (2015).
Zhao, L. et al. Lanosterol reverses protein aggregation in cataracts. Nature 523, 607–611 (2015).
Andley, U. P., Tycksen, E., McGlasson-Naumann, B. N. & Hamilton, P. D. Probing the changes in gene expression due to α-crystallin mutations in mouse models of hereditary human cataract. PLoS ONE 13, e0190817 (2018).
Koontz, L. TCA precipitation. Methods Enzymol. 541, 3–10 (2014).
Natale, M., Maresca, B., Abrescia, P. & Bucci, E. M. Image analysis workflow for 2-D electrophoresis gels based on ImageJ. Proteom. Insights 4, 37 (2011).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Preis, W., Bestehorn, A., Buchner, J. & Haslbeck, M. An alternative splice variant of human αA-crystallin modulates the oligomer ensemble and the chaperone activity of α-crystallins. Cell Stress Chaperones 22, 541–552 (2017).
Courchesne, P. L. & Patterson, S. D. Identification of proteins by matrix-assisted laser desorption/ionization mass spectrometry using peptide and fragment ion masses. Methods Mol. Biol. 112, 487–511 (1999).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Dulbecco, R. & Vogt, M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 99, 167–182 (1954).
Jeong, J. Y. et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 78, 5440–5443 (2012).
Philo, J. S. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 354, 238–246 (2006).
Stafford, W. F. III. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 203, 295–301 (1992).
Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science. (eds. Harding, S. E. et al.) 90−125 (The Royal Society of Chemistry, 1992).
Acknowledgements
We thank D. Catici for experimental help; E. Bürkle, M. Stadler, G.-M. Feind and F. Rührnößl for their technical assistance; as well as F. Tippel and M. Schwieder from NanoTemper Technologies GmbH for the kind support with our nanoDSF measurements. This work was supported by a grant from the DFG (SFB 1035) to S.W., M.H. and J.B. This research was further supported by the Austrian Science Foundation (P28854, I3792, DK-MCD W1226 to T.M.), the Austrian Research Promotion Agency (FFG: 864690, 870454), the Integrative Metabolism Research Center Graz, the Austrian infrastructure program 2016/2017, the Styrian government (Zukunftsfonds) and BioTechMed/Graz. N.C.H.L. acknowledges funding from the Alexander von Humboldt Foundation. We thank H. Ehmann, J. Gautsch, P. Kotnik and A. Scheiflinger-Latal from Anton Paar Graz for their technical support with SAXS experiments on intact eye lenses and the production of a sample holder for eye lenses, as well as M. Kriechbaum for SAXS support. We thank F.-X. Schmid for comments on the manuscript and discussions.
Author information
Authors and Affiliations
Contributions
P.W.N.S, N.C.H.L, C.D., O.P., O.V.A., M.H., S.W., J.G, T.M. and J.B. designed the study, interpreted data and wrote the manuscript. P.W.N.S, N.C.H.L, C.P., K.C.B, B.R., B.B., F.P., J.P. and T.M. performed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Roy Quinlan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of the proteome of wild-type mice.
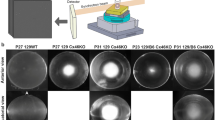
(a) Graphs show different measurements from C3HeB/FeJ (squares) and C57Bl/6 J (circles) mice. Top, body weight (filled symbols) and lens wet weight (open symbols). Middle and bottom, amounts of extractable water-soluble and -insoluble protein per lens wet weight was determined by the Bradford protein assay, using BSA as a standard. Data are given as mean and s.d. per lens pair (left and right eye) of one animal. Please see Supplementary Table 2 and 3 for exact sample size. (b) 2-DE map of the water-soluble fraction of lenses from newborn wild-type mice. For densitometric analysis (n = 2 individual animals), only the low molecular crystallin region was considered (see excerpt). The fraction of α- (gold), β- (orange) and γ-crystallins (blue) was determined up to 12 months of age, for the water-soluble (solid bars) and -insoluble (shaded bars) fraction. Please see Supplementary Data 1 for numerical values. Scheimpflug images of C57Bl/6 J lenses at 3, 6 and 12 months are shown as a reference for wild-type behavior.
Extended Data Fig. 2 Changes in the proportion of crystallin isoforms during aging in wild-type and mutant mice.
Graphs show relative proportions of single crystallin isoforms, plotted against age, in the water-soluble or -insoluble fractions from whole lens extracts (C3HeB/FeJ (black squares), αA mutant (red circles), γD mutant (red triangles) and C57Bl/6 J (black squares), βA2 mutant (red circles)). Data are shown as mean and s.d. for n = 2 individual animals. Data behind graphs are in Supplementary Data 1. The sum of the isoform proportions equals 1 and the analyses were performed for the water-soluble and water-insoluble fraction.
Extended Data Fig. 3 Characterization of the protein complexes found in the lens extracts from wild-type mice.
(a) Top, SEC-HPLC profiles of water-soluble proteins in whole lens extracts for both wild-type strains. The elution profiles of samples from two mice (pair of lenses, each) are shown (as solid and traced lines). Bottom, the integrated signal was plotted against the age to follow fractional changes over time; fractions are color-coded as shown on top. Data are means and s.d. from the 2 replicates shown on top. (b) Top, 2-DE and EM analyses to check for the protein composition of isolated HMW and αL fraction. Scale bar: 50 nm. Bottom, crystallin proportions were determined by densitometric analysis of the 2-DE gels (light grey: αA-crystallin, dark grey: αB-crystallin, orange: β-crystallins, blue: γ-crystallins). (c) SV-AUC profiles showing analyses of αL particles from two mice each, revealing a slight increase in their sedimentation coefficient (s(20,w)) during aging (grey and black lines). Error bars result from averaging consecutive scans during SV-AUC experiments. Apparent maxima of the g(s*) distribution show the most populated particle species. (d) Scattering profiles and Guinier plots of lens extracts from C3HeB/FeJ wild-type mice at different ages: 1 (black), 3 (grey) and 9 (red) months.
Extended Data Fig. 4 2-DE of crystallin fractions from wild-type and mutant mice.
After separation by SEC-HPLC, the individual fractions were checked for their protein composition using 2-DE. Note that the type of ampholyte used (Pharmalyte) caused a staining artifact but that did not influence IEF or second dimension separation of the proteins, as shown at the bottom right. For densitometric analysis, the background signal of 2-DE gels was subtracted using the rolling ball background correction with 100 px in ImageJ. For each individual fraction, one 2-DE experiment was performed.
Extended Data Fig. 5 Cataract-associated crystallin mutants are thermodynamically destabilized.
(a) Far-UV CD spectroscopy analyses of secondary structure analysis of recombinant wild-type (black) and mutant (red) crystallins. Due to low solubility, no spectra were recorded for βA2-S47P. (b) NanoDSF measurements, recording the optical density to detect protein aggregation. The V124E mutation did not change the aggregation propensity of αA-crystallin, but decreased the chaperone activity towards the model substrate L-MDH (inset). Data shown as mean and s.d. from n = 3 independent samples, obtained with recombinant protein from the same batch. Wild-type crystallin (black) and crystallin mutant (red). Experiments were carried out in PBS, but for the βA2 mutant. Due to the low stability of βA2 S47P, Tris buffer was used and L-arginine was added for the measurements. The measurement with wild-type βA2 in PBS are shown in black and in grey for the Tris/Arginine buffer as a reference (c) SEC-MALS/ -HPLC and SV-AUC (inset) were employed to characterize the quaternary structure of wild-type (black) and mutant (red) crystallins. Note: Due to the low stability of βA2-S47P, quantitative unfolding of the protein occurs during SV-AUC runs. Data shown resembles a representative distribution from three independent samples, obtained with recombinant protein from the same batch. Error bars result from averaging consecutive scans during SV-AUC experiments. Apparent maxima of the g(s*) distribution show the most populated particle species.
Extended Data Fig. 6 Characterization of the proteome of mutant mice.
(a) Left, body weight (filled symbols) and lens wet weight (open symbols), per pair of eye lenses from one individual: αA mutant (squares), βA2 mutant (circles) and γD mutant (triangles). Middle and right, amount of water-soluble and -insoluble protein was determined by the Bradford protein assay using BSA as a standard. Data are given as mean ± SD of biological replicates (animals or pairs of eye lenses; please see Supplementary Table 2 and 3 for exact sample size). (b) Top, 2-DE map of the water-soluble fraction of lenses from newborn wild-type mice. Bottom, densitometric analysis; only the low molecular crystallin region was considered (see excerpt). The fraction of α- (gold), β- (orange) and γ-crystallins (blue) was determined up to 12 months of age, for the water-soluble (solid bars) and -insoluble (shaded bars) fraction. Please see Supplementary Data 1 for numerical values.
Extended Data Fig. 7 Comparison of extraction efficiency using different buffers.
2-DE maps showing comparison of the eye lens proteome pattern obtained for the water-insoluble fraction of the three mutants studied, solubilized in the presence of 6 M urea or 7 M urea plus 2 M thiourea. One experiment was performed per mutant, age and extraction buffer.
Extended Data Fig. 8 Protein distribution in whole lens extracts.
(a) The proportion of α-, β- and γ-crystallins was plotted as X/Y/Z-coordinates for the water-soluble and -insoluble fraction, to illustrate the distribution of crystallins in whole lens extracts. Data shown as mean and s.d. for n = 2; data from Supplementary Data 1. The sum of the three crystallin families equals 100 % with α-crystallins representing the Y-coordinate, β-crystallins the X-coordinate and γ-crystallins the Z-coordinate. The composition is changed in the mutants and EMORY mice when cataract is present. At 1 month, when the lens of EMORY mice is still clear, the crystallin composition is very similar to the wild-type composition. During aging, the γ-crystallins gradually decrease (Z-axis). (C57Bl/6 J (black circles), C3HeB/FeJ (black squares), αA mutant (red squares), βA2 mutant (red circles), γD mutant (red triangles), βB2 mutant (green circles), and EMORY (blue circles)). (b) The ratio of α-/β-crystallin content is changed in mutants for both fractions (C3HeB/FeJ (black squares), C57Bl/6 J (black circles), αA mutant (red squares), γD mutant (red circles), and βA2 mutant (red triangles)). Data shown are mean and s.d. for n = 2 animals (pairs of eye lenses).
Supplementary information
Supplementary Information
Supplementary Fig. 1, Supplementary Tables 1–5 and Supplementary Note 1.
Supplementary Data 1
Input data for lens extract analyses used in Fig. 1, Fig. 3, ED Fig. 1, ED Fig. 2, ED Fig. 3 and ED Fig. 6
Rights and permissions
About this article
Cite this article
Schmid, P.W.N., Lim, N.C.H., Peters, C. et al. Imbalances in the eye lens proteome are linked to cataract formation. Nat Struct Mol Biol 28, 143–151 (2021). https://doi.org/10.1038/s41594-020-00543-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-00543-9
This article is cited by
-
Structural biology in the Anthropocene epoch
Nature Structural & Molecular Biology (2024)
-
Quantitative X-ray tomographic analysis reveals calcium precipitation in cataractogenesis
Scientific Reports (2021)