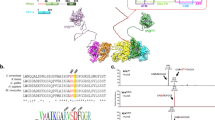
Abstract
In response to DNA damage or replication fork stalling, the basal activity of Mec1ATR is stimulated in a cell-cycle-dependent manner, leading to cell-cycle arrest and the promotion of DNA repair. Mec1ATR dysfunction leads to cell death in yeast and causes chromosome instability and embryonic lethality in mammals. Thus, ATR is a major target for cancer therapies in homologous recombination–deficient cancers. Here we identify a single mutation in Mec1, conserved in ATR, that results in constitutive activity. Using cryo-electron microscopy, we determine the structures of this constitutively active form (Mec1(F2244L)-Ddc2) at 2.8 Å and the wild type at 3.8 Å, both in complex with Mg2+-AMP-PNP. These structures yield a near-complete atomic model for Mec1–Ddc2 and uncover the molecular basis for low basal activity and the conformational changes required for activation. Combined with biochemical and genetic data, we discover key regulatory regions and propose a Mec1 activation mechanism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The cryo-EM reconstruction volumes and the atomic coordinates generated in this study are available at the EMDB under accession codes EMD-11050 (nucleotide-bound F2244L mutant State I), EMD-11051 (nucleotide-bound F2244L mutant State II), EMD-11055 (nucleotide-bound wild type) and EMD-11056 (wild type); and the RCSB Protein Data Bank under the PDB codes 6Z2W (AMP-PNP-bound F2244L State I), 6Z2X (AMP-PNP-bound F2244L State II) and 6Z3A (AMP-PNP-bound wild type). Yeast strains, plasmids and plasmid sequences are available upon request. Source data are provided with this paper.
References
Weber, A. M. & Ryan, A. J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 149, 124–138 (2015).
Lovejoy, C. A. & Cortez, D. Common mechanisms of PIKK regulation. DNA Repair (Amst.) 8, 1004–1008 (2009).
Kumagai, A., Lee, J., Yoo, H. Y. & Dunphy, W. G. TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 (2006).
Mordes, D. A., Nam, E. A. & Cortez, D. Dpb11 activates the Mec1–Ddc2 complex. Proc. Natl Acad. Sci. USA 105, 18730–18734 (2008).
Mordes, D. A., Glick, G. G., Zhao, R. & Cortez, D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22, 1478–1489 (2008).
Navadgi-Patil, V. M. & Burgers, P. M. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 283, 35853–35859 (2008).
Navadgi-Patil, V. M. & Burgers, P. M. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol. Cell 36, 743–753 (2009).
Kumar, S. & Burgers, P. M. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 27, 313–321 (2013).
Paull, T. T. Mechanisms of ATM activation. Annu. Rev. Biochem. 84, 711–738 (2015).
Hailemariam, S., Kumar, S. & Burgers, P. M. Activation of Tel1ATM kinase requires Rad50 ATPase and long nucleosome-free DNA but no DNA ends. J. Biol. Chem. 294, 10120–10130 (2019).
Wanrooij, P. H., Tannous, E., Kumar, S., Navadgi-Patil, V. M. & Burgers, P. M. Probing the Mec1ATR checkpoint activation mechanism with small peptides. J. Biol. Chem. 291, 393–401 (2016).
Thada, V. & Cortez, D. Common motifs in ETAA1 and TOPBP1 required for ATR kinase activation. J. Biol. Chem. 294, 8395–8402 (2019).
Sawicka, M. et al. The dimeric architecture of checkpoint kinases Mec1ATR and Tel1ATM reveal a common structural organization. J. Biol. Chem. 291, 13436–13447 (2016).
Ball, H. L. & Cortez, D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J. Biol. Chem. 280, 31390–31396 (2005).
Deshpande, I. et al. Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage. Mol. Cell 68, 431–445.e5 (2017).
Zou, L. & Elledge, S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003).
Memisoglu, G. et al. Mec1ATR autophosphorylation and Ddc2ATRIP phosphorylation regulates DNA damage checkpoint signaling. Cell Rep. 28, 1090–1102.e3 (2019).
Adams, J. A. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101, 2271–2290 (2001).
Lempiainen, H. & Halazonetis, T. D. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 28, 3067–3073 (2009).
Yang, H. et al. mTOR kinase structure, mechanism and regulation. Nature 497, 217–223 (2013).
Yates, L. A. et al. Cryo-EM structure of nucleotide-bound Tel1ATM unravels the molecular basis of inhibition and structural rationale for disease-associated mutations. Structure 28, 96–104.e3 (2020).
Sibanda, B. L., Chirgadze, D. Y., Ascher, D. B. & Blundell, T. L. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 355, 520–524 (2017).
Gat, Y. et al. InsP6 binding to PIKK kinases revealed by the cryo-EM structure of an SMG1–SMG8–SMG9 complex. Nat. Struct. Mol. Biol. 26, 1089–1093 (2019).
Jansma, M. et al. Near-complete structure and model of Tel1ATM from Chaetomium thermophilum reveals a robust autoinhibited ATP state. Structure 28, 83–95.e5 (2020).
Mallory, J. C. & Petes, T. D. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl Acad. Sci. USA 97, 13749–13754 (2000).
Paciotti, V., Clerici, M., Scotti, M., Lucchini, G. & Longhese, M. P. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21, 3913–3925 (2001).
Wang, X. et al. 3.9 Å structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP. Science 358, 1206–1209 (2017).
Rao, Q. et al. Cryo-EM structure of human ATR-ATRIP complex. Cell Res. 28, 143–156 (2018).
Ball, H. L. et al. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 27, 3367–3377 (2007).
Gangadhara, G. et al. A class of highly selective inhibitors bind to an active state of PI3Kγ. Nat. Chem. Biol. 15, 348–357 (2019).
Wanrooij, P. H. & Burgers, P. M. Yet another job for Dna2: checkpoint activation. DNA Repair (Amst.) 32, 17–23 (2015).
Jacobsen, D. M., Bao, Z. Q., O’Brien, P., Brooks, C. L. III. & Young, M. A. Price to be paid for two-metal catalysis: magnesium ions that accelerate chemistry unavoidably limit product release from a protein kinase. J. Am. Chem. Soc. 134, 15357–15370 (2012).
Burtelow, M. A., Roos-Mattjus, P. M., Rauen, M., Babendure, J. R. & Karnitz, L. M. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J. Biol. Chem. 276, 25903–25909 (2001).
Majka, J. & Burgers, P. M. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl Acad. Sci. USA 100, 2249–2254 (2003).
Allen, J. B., Zhou, Z., Siede, W., Friedberg, E. C. & Elledge, S. J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8, 2401–2415 (1994).
Sanchez, Y. et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357–360 (1996).
Ma, J. L., Lee, S. J., Duong, J. K. & Stern, D. F. Activation of the checkpoint kinase Rad53 by the phosphatidyl inositol kinase-like kinase Mec1. J. Biol. Chem. 281, 3954–3963 (2006).
Downs, J. A., Lowndes, N. F. & Jackson, S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).
Fink, M., Imholz, D. & Thoma, F. Contribution of the serine 129 of histone H2A to chromatin structure. Mol. Cell. Biol. 27, 3589–3600 (2007).
Puddu, F., Piergiovanni, G., Plevani, P. & Muzi-Falconi, M. Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase ε. PLoS Genet. 7, e1002022 (2011).
Lanz, M. C. et al. Separable roles for Mec1/ATR in genome maintenance, DNA replication, and checkpoint signaling. Genes Dev. 32, 822–835 (2018).
Bandhu, A., Kang, J., Fukunaga, K., Goto, G. & Sugimoto, K. Ddc2 mediates Mec1 activation through a Ddc1- or Dpb11-independent mechanism. PLoS Genet. 10, e1004136 (2014).
Rouse, J. & Jackson, S. P. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9, 857–869 (2002).
Huse, M. & Kuriyan, J. The conformational plasticity of protein kinases. Cell 109, 275–282 (2002).
Bao, Z. Q., Jacobsen, D. M. & Young, M. A. Briefly bound to activate: transient binding of a second catalytic magnesium activates the structure and dynamics of CDK2 kinase for catalysis. Structure 19, 675–690 (2011).
Williams, R. M., Yates, L. A. & Zhang, X. Structures and regulations of ATM and ATR, master kinases in genome integrity. Curr. Opin. Struct. Biol. 61, 98–105 (2020).
Ung, P. M. & Schlessinger, A. DFGmodel: predicting protein kinase structures in inactive states for structure-based discovery of type-II inhibitors. ACS Chem. Biol. 10, 269–278 (2015).
Modi, V. & Dunbrack, R. L. Jr. Defining a new nomenclature for the structures of active and inactive kinases. Proc. Natl Acad. Sci. USA 116, 6818–6827 (2019).
Jauch, R. et al. Mitogen-activated protein kinases interacting kinases are autoinhibited by a reprogrammed activation segment. EMBO J. 25, 4020–4032 (2006).
Yang, H. et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 (2017).
Yin, X., Liu, M., Tian, Y., Wang, J. & Xu, Y. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 27, 1341–1350 (2017).
Bastidas, A. C. et al. Phosphoryl transfer by protein kinase A is captured in a crystal lattice. J. Am. Chem. Soc. 135, 4788–4798 (2013).
McDonald, J. P., Levine, A. S. & Woodgate, R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147, 1557–1568 (1997).
Zhao, X., Muller, E. G. & Rothstein, R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2, 329–340 (1998).
Chabes, A., Domkin, V. & Thelander, L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274, 36679–36683 (1999).
Hailemariam, S. et al. The telomere-binding protein Rif2 and ATP-bound Rad50 have opposing roles in the activation of yeast Tel1ATM kinase. J. Biol. Chem. 294, 18846–18852 (2019).
Hustedt, N. & Shimada, K. Analyzing DNA replication checkpoint in budding yeast. Methods Mol. Biol. 1170, 321–341 (2014).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. Elife 7, e35383 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Burnley, T., Palmer, C. M. & Winn, M. Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 73, 469–477 (2017).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B., Wedell, J. R., Wenger, R. K., Ulrich, E. L. & Markley, J. L. MolProbity for the masses—of data. J. Biomol. NMR 63, 77–83 (2015).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Acknowledgements
We thank Burgers laboratory members C. Stith and B. Yoder for strains construction, and J. Haber (Brandeis University) for plasmids. We thank A. Nan (Francis Crick Institute), K. Cunnea (eBIC) and Y. Song (eBIC) for their support with cryo-EM data acquisition and Zhang laboratory members R. Ayala, for help with initial screening, and R. Williams, for discussions. Initial cryo-EM screening of samples was carried out at the Imperial College London Center for Structural Biology EM facility. High resolution cryo-EM data were collected at eBIC (proposal no. EM19865). eBIC is funded by the Wellcome Trust, MRC and BBSRC. This work was funded in part by grant no. GM118129 from the National Institutes of Health (to P.M.B.) and the Wellcome Trust grant no. 210658/Z/18/Z (to X.Z.).
Author information
Authors and Affiliations
Contributions
E.A.T., L.A.Y., X.Z. and P.M.B. planned this study. E.A.T. carried out the biochemical and genetic studies. L.A.Y. carried out the structural studies. All authors were involved in the interpretation of the results and the writing of the paper and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Peer reviewer reports are available. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
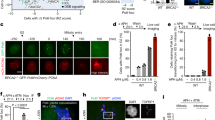
Extended Data Fig. 1 Activation loop mutagenesis of Mec1.
a, Representative complete gel of Mec1 kinase assay, described in Fig. 2. The gel shown is for the experiment in Fig. 2a, WT. b, Kinase activity of Mec1 and Mec1(F2244L) as a function of Dna2(1-499). Phosphorylation rates are given below the gel. c, Kinase activity of Mec1 and Mec1(F2244L) as a function of Dna2-1 peptide: HHDFTQDEDGPMEEVIWKYSPLQRDMSDKT. Fold stimulation compared to wild-type Mec1 without activator is given. d, Phylogenetic analysis of the activation loop 2243DFD2245 motif. 640 eukaryotic Mec1/ATR sequences were aligned with MSAProbs (https://toolkit.tuebingen.mpg.de/), filtered to a set of 95 sequences that showed less than 50% sequence identity, and the motif distribution recorded. e, Titration of Rad53 into the Mec1 assay. Standard assays with 3 nM Mec1 and 5 nM Dna2(1-499) activator, or with 3 nM Mec1(F2244L) with or without 5 nM Dna2(1-499) activator were carried out at increasing concentrations of GST-Rad53-kd. Activities are expressed as Rad53 phosphates per Mec1 (monomer) per minute, and the data were modeled to the Michaelis-Menten equation. f, Ponceau staining of the extracts used for the Western blots in Fig. 2e.
Extended Data Fig. 2 Cell cycle analysis of a MEC1 mutant with constitutive activity.
a, Western blot analysis of phospho-H2A (pS129) (top) and Rad53 (bottom) in wild-type strain PY405, with either MEC1 or mec1-F2244L. Cells were arrested in G1 phase with alpha-factor, or arrested in G1 phase with alpha-factor, and released into S phase with 200 mM of hydroxyurea for the indicated time. Ponceau staining of the blot is shown below. b, mec1-F2244L suppresses the growth defect of activator-defective yeast. In the experimental scheme, the extreme defects of the activator-defective strain were initially suppressed by a plasmid-borne copy of wild-type DNA2, containing the URA3 gene as selectable and counterselectable marker. Thus, strain MEC1 tel1Δ ddc1Δ dna2Δ (PY270) contains three plasmids: p(DNA2 URA3), p(dna2-WYAA TRP1), and either vector or p(mec1-x LEU2). The strains were grown on media lacking Trp and Leu (left), or on 5FOA-containing media (right) that only permits growth if the p(DNA2 URA3) plasmid is lost. The data indicate that mec1-F2244L allows cell growth without p(DNA2 URA3), therefore suppressing the growth defect of the activator-defective strain. c, Ponceau staining of the extracts used for the blots in Fig. 2g. d, Constitutively active mec1-F2244L progresses slowly through S phase. Strain PY406 containing p(MEC1 LEU2) (blue) or p(mec1-F2244L LEU2) (red). Cell cycle distribution was measured for (a) asynchronous cells; (b) alpha-factor arrested G1 cells; (c) G1 arrested cells treated with 4NQO for 30 min; (d, e, f) G1 arrested cells released into fresh YPD for 5, 30, and 60 minutes; (g) G1 arrested cells released into fresh YPD containing 200 mM hydroxyurea for 60 minutes, (h, i, j) example of gating strategy shown for plot (a) p(MEC1 LEU2) asynchronous cells.
Extended Data Fig. 3 CryoEM processing and reconstruction quality of Mec1-Ddc2.
a, b, Processing tree resulting reconstructions of Mec1-Ddc2 and a second reconstruction in complex with AMP-PNP (see Methods for details). c, f, Local resolution estimates from ResMap, with slice through the density to show internal features, of apo (c) and bound with AMP-PNP (f). d, g, Angular distribution from CisTEM autorefine, and (e, h) Gold-standard Fourier shell correlation (FSC) from RELION-3.0 and CisTEM.
Extended Data Fig. 4 Map and model features of the Mec1-Ddc2 complex.
a, 2D classes of Mec1-Ddc2 after a focused 3D refinement masking on Mec1-Ddc2 heterodimer, showing intrinsic flexibility of the complex across the dimer interface. b, Electron density features of the bound AMP-PNP, and c, strong electron density (unsharpened map) showing the PRD-I interaction with the activation loop at two points (asterisked). d, Map to model FSC curves.
Extended Data Fig. 5 CryoEM processing and reconstruction quality of Mec1(F2244L)-Ddc2.
a, Processing tree resulting in high resolution reconstructions of Mec1(F2244L)-Ddc2 in complex with AMP-PNP and magnesium captured in two states (see Methods). b, Local resolution estimates from ResMap, with slice through the density to show internal features, of State I, (c) angular distribution from CisTEM autorefine, and (d) Gold-standard Fourier shell correlation (FSC) from RELION-3.0 and CisTEM. e, Local resolution estimates from ResMap of State II, with (f) angular distribution from CisTEM autorefine, and (g) Gold-standard Fourier shell correlation (FSC) from RELION-3.0 and CisTEM.
Extended Data Fig. 6 Data and model quality of the Mec1(F2244L)-Ddc2 reconstruction.
a, Map to model FSC curves of the F2244 mutant reconstruction. b, Ddc2 and Mec1 N-terminal domain density (NTD) and model showing clear separation of Mec1 and Ddc2 proteins for accurate model building of this region (left), with close-up views of chain tracing between the model built in this study (middle) and the previously published model (PDB:5X6O) (right). The arrow indicates the point at which the two models diverge. c, High-resolution features from the 2.8 Å map, showing that the electron density quality is sufficient to resolve types of aromatic residues (Phenylalanine over Tyrosines), β−branched side chains (Isoleucine), as well as smaller hydrophobics (Valine) and an example of a split conformation of Arginine. d-f, CryoEM density regions of the Mec1 N-terminal domain (~300 amino acids) showing the overall fit of the model and side chains (labeled), along with close-up views of different regions showing unambiguous side chain density for accurate model building.
Extended Data Fig. 7 Global and kinase domain structural comparisons of Mec1-Ddc2.
a-d, Structural comparisons between the Mec1 model (a,b) and Ddc2 model (c,d) from this study and the PDB:5X6O showing the global differences in N-terminal domains of both proteins. e-g, Overall comparison between the kinase region of Mec1(F2244L) State I (gray) and State II (light gray), demonstrating that both states are very similar outside of the active site with an Rmsd = 0.3 Å. f-g, Electron density of the nucleotide binding site and different side chain conformations associated with State I and State II (see main text for details).
Extended Data Fig. 8 ATP dependence of Mec1 activity.
a, Standard Mec1 kinase assays without activator at 40 mM NaCl, or (b) with 200 nM Dna2(1-499) at 100 mM NaCl, were carried out at increasing concentrations of ATP. Activities are expressed as protein phosphates (Rad53 plus Dna2(1-499) when relevant) per Mec1 (monomer) per minute. c, Comparative ATPase (solid bars) and kinase (striped bars) activities of Mec1-Ddc2 and Mec1(F2244L)-Ddc2, in the presence or absence of Rad53 and Dpb11 (see Methods). d, Summary of phenotypes of all Mec1 mutants. The in vitro and in vivo phenotypes of the mutants are shown in the form of heat maps using Prism 8 GraphPad software.
Extended Data Fig. 9 Structural analysis and comparisons of Mec1.
a-c, Electron density of the activation loop from the apo (a), AMP-PNP-bound (b) and AMP-PNP-bound F2244L mutant (c), showing that in all cases the activation loop remains ordered, with flexibility around the DFD-motif (asterisks), which could not be easily resolved in the wild-type structures. Several large residues are shown as landmarks. d,e, PRD-I hydrophobic network comparisons between Mec1-Ddc2 (e) and Tel1 (e), suggesting that M2312 plays an analogous role to W2701 in Tel1. PRD-I, activation loop and catalytic loop are colored as in Fig. 1b. f-i, Comparison of stabilizing interactions in activations loops across PIKKs. f, In Mec1, the DFD+1 residue plays a role in stabilizing the activation loop. In our activated structure the thiol group of the invariant C2246 forms an H-bond with the main chain carbonyl of L2222 of the catalytic loop, helping to stabilize the active state. In Tel1 the DLG+1 (I2634) forms a hydrophobic spline with G2639 and L2642 of the activation loop (g), whereas in mTOR (h) and DNA-PKcs (i) an ion pair is preferred.
Supplementary information
Supplementary Video 1
Global motion of the Mec1–Ddc2 complex upon activation. The video shows conformational changes across the Mec1–Ddc2 complex dimer interface when aligned on a single protomer. The trajectories between structures were calculated and visualized by morphing between AMP-PNP-bound wild-type auto-inhibited Mec1–Ddc2 and the AMP-PNP-bound constitutively active Mec1(F2244L)–Ddc2 mutant structure. Domains are colored as in Fig. 2.
Supplementary Video 2
Kinase domain motion upon activation. Conformational changes in the Mec1 kinase domain visualized by morphing between the AMP-PNP-bound wild-type Mec1 auto-inhibited state and the AMP-PNP-bound Mec1(F2244L) constitutively active mutant state. Kinase domain features are colored as in Fig. 4f.
Supplementary Video 3
Molecular details of PRD-I retraction. Conformational changes in the kinase active site visualized by morphing between wild-type auto-inhibited and Mec1(F2244L) constitutively active State I, with further motion to State II, all captured by cryo-EM in this study. Kinase domain features are colored as in Fig. 7.
Source data
Source Data Fig. 2
Unprocessed gel images and western blots
Source Data Fig. 2
Statistical data for graphs
Source Data Fig. 6
Statistical data for graphs
Source Data Fig. 7
Statistical data for graphs
Rights and permissions
About this article
Cite this article
Tannous, E.A., Yates, L.A., Zhang, X. et al. Mechanism of auto-inhibition and activation of Mec1ATR checkpoint kinase. Nat Struct Mol Biol 28, 50–61 (2021). https://doi.org/10.1038/s41594-020-00522-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-00522-0
This article is cited by
-
DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp
Nature Structural & Molecular Biology (2022)
-
Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins
Cell Discovery (2022)
-
Molecular basis of human ATM kinase inhibition
Nature Structural & Molecular Biology (2021)
-
The activation mechanisms of master kinases in the DNA damage response
Genome Instability & Disease (2021)