Abstract
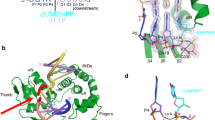
DNA double-strand breaks are the most dangerous type of DNA damage and, if not repaired correctly, can lead to cancer. In humans, Ku70/80 recognizes DNA broken ends and recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form DNA-dependent protein kinase holoenzyme (DNA-PK) in the process of non-homologous end joining (NHEJ). We present a 2.8-Å-resolution cryo-EM structure of DNA-PKcs, allowing precise amino acid sequence registration in regions uninterpreted in previous 4.3-Å X-ray maps. We also report a cryo-EM structure of DNA-PK at 3.5-Å resolution and reveal a dimer mediated by the Ku80 C terminus. Central to dimer formation is a domain swap of the conserved C-terminal helix of Ku80. Our results suggest a new mechanism for NHEJ utilizing a DNA-PK dimer to bring broken DNA ends together. Furthermore, drug inhibition of NHEJ in combination with chemo- and radiotherapy has proved successful, making these models central to structure-based drug targeting efforts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data supporting the findings of this paper are available from the corresponding author upon reasonable request. All data generated or analysed during this study are included in this published Article and its Supplementary Information files. Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-11211, 11185, 11213, 11217, 11216, 11219 and 11215. Atomic coordinates have been deposited in the RCSB Protein Data Bank under accession codes PDB 6ZH2, 6ZFP, 6ZH4, 6ZHA, 6ZH8, 6ZHE and 6ZH6. Source data are provided with this paper.
References
Chirgadze, D. Y., Ascher, D. B., Blundell, T. L. & Sibanda, B. L. DNA-PKcs, allostery and DNA double-strand break repair: defining the structure and setting the stage. Methods Enzymol. 592, 145–157 (2017).
Liang, S. et al. Achieving selectivity in space and time with DNA double-strand-break response and repair: molecular stages and scaffolds come with strings attached. Stuct. Chem. 28, 161–171 (2017).
Walker, J. R., Corpina, R. A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001).
Gell, D. & Jackson, S. P. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 27, 3494–3502 (1999).
Singleton, B. K., Torres-Arzayus, M. I., Rottinghaus, S. T., Taccioli, G. E. & Jeggo, P. A. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 19, 3267–3277 (1999).
Meek, K., Dang, V. & Lees-Miller, S. P. DNA-PK: the means to justify the ends? Adv. Immunol. 99, 33–58 (2008).
Smith, G. C. & Jackson, S. P. The DNA-dependent protein kinase. Genes Dev. 13, 916–934 (1999).
Carter, T., Vancurova, I., Sun, I., Lou, W. & DeLeon, S. A DNA-activated protein kinase from HeLa cell nuclei. Mol. Cell. Biol. 10, 6460–6471 (1990).
Sibanda, B. L., Chirgadze, D. Y. & Blundell, T. L. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463, 118–121 (2010).
Hammel, M. et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 285, 1414–1423 (2010).
Jiang, W. et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell 58, 172–185 (2015).
Sibanda, B. L., Chirgadze, D. Y., Ascher, D. B. & Blundell, T. L. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 355, 520–524 (2017).
Yin, X., Liu, M., Tian, Y., Wang, J. & Xu, Y. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 27, 1341–1350 (2017).
Terwilliger, T. L., Ludtke, S. J., Read, R. R., Adams, P. D. & Afonine, P. V. Improvement of cryo-EM maps by density modification. Nat. Methods 17, 923–927 (2020).
Sharif, H. et al. Cryo-EM structure of the DNA-PK holoenzyme. Proc. Natl Acad. Sci. USA 114, 7367–7372 (2017).
Cui, X. et al. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 25, 10842–10852 (2005).
Kidmose, R. T. et al. Namdinator—automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Zhang, Z. et al. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein–protein interactions. Structure 12, 495–502 (2004).
Falck, J., Coates, J. & Jackson, S. P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 (2005).
Spagnolo, L., Rivera-Calzada, A., Pearl, L. H. & Llorca, O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519 (2006).
Brouwer, I. et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–569 (2016).
Ropars, V. et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl Acad. Sci. USA 108, 12663–12668 (2011).
Hammel, M. et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 286, 32638–32650 (2011).
Wu, Q. et al. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem. Soc. Trans. 39, 1387–1392 (2011).
Andres, S. N. et al. A human XRCC4–XLF complex bridges DNA. Nucleic Acids Res. 40, 1868–1878 (2012).
Ahnesorg, P., Smith, P. & Jackson, S. P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124, 301–313 (2006).
Mahaney, B. L., Hammel, M., Meek, K., Tainer, J. A. & Lees-Miller, S. P. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem. Cell Biol. 91, 31–41 (2013).
Hammel, M. et al. An intrinsically disordered APLF links Ku, DNA-PKcs and XRCC4-DNA ligase IV in an extended flexible non-homologous end joining complex. J. Biol. Chem. 291, 26987–27006 (2016).
Chen, X. et al. Cutting antiparallel DNA strands in a single active site. Nat. Struct. Mol. Biol. 27, 119–126 (2020).
Gellert, M. V(D)J recombination: RAG proteins, repair factors and regulation. Annu. Rev. Biochem. 71, 101–132 (2002).
Kim, M. S., Lapkouski, M., Yang, W. & Gellert, M. Crystal structure of the V(D)J recombinase RAG1–RAG2. Nature 518, 507–511 (2015).
Schatz, D. G. & Swanson, P. C. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45, 167–202 (2011).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Punjani, A., Brubaker, M. A. & Fleet, D. J. Building proteins in a day: efficient 3D molecular structure estimation with electron cryomicroscopy. IEEE Trans. Pattern Anal. Mach. Intell. 39, 706–718 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Acknowledgements
Model coordinates have been deposited in the Protein Data Bank and maps deposited at the Electron Microscopy Data Bank under the accession numbers shown in Table 1. We thank T. Terwilliger for advice regarding the usage of the ResolveCryoEM program in Phenix. We also thank B. Luisi, L. Pellegrini and N. Rzechorzek for their critical reading and feedback on the manuscript. We are grateful to the Wellcome Trust for a Program Grant (O93167/Z/10/Z; 2011–2016) and Investigator Award (200814/Z/16/Z; 2016) to support this research.
Author information
Authors and Affiliations
Contributions
A.K.C. purified and prepared complexes for cryo-EM, modelled the protein structures and wrote the manuscript. S.W.H. collected and processed the cryo-EM data and assisted with modelling the protein structures and writing the manuscript. S.L. carried out the initial cryo-EM analysis of DNA-PKcs. A.K.S. assisted with Ku70/80 expression and purification. A.H. expressed and purified the Ku80 CTR. D.Y.C. collected cryo-EM data and provided expertise. L.R.C. assisted with grid preparation and collected cryo-EM data. T.M.D.O. helped with research design and advice. T.L.B. directed the study, provided advice and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
T.M.D.O. is employed at AstraZeneca. The authors declare no other competing interests.
Additional information
Peer review information Peer reviewer reports are available. Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 DNA-PK complex formation and optimisation of cryo-EM samples.
a, Gel filtration profile of DNA-PKcs, Ku70/80 and the DNA-PK complex with Y-shaped 42-55 bp DNA. Inset, SDS-PAGE gel for DNA-PKcs, Ku70/80 and DNA-PK with molecular weight marker (kDa) indicated. Uncropped SDS gel image is available as Source Data. b and c, Example of 2D classes of DNA-PK and the angular distribution calculated in cryoSPARC for particle projections shown as a heat map, where b is before addition of CHAPSO and c after CHAPSO has been added. Scale bar = 20nm.
Extended Data Fig. 2 Cryo-EM maps before and after using ResolveCryoEM.
a, DNA-PKcs (state 2), b, DNA-PK monomer and c, DNA-PK dimer before (as green sticks) and after ResolveCryoEM (yellow sticks) for residues 3897-391814.
Extended Data Fig. 3 Sequence of DNA-PKcs.
Coloured blocks below the sequence represent a traffic light comparison to the modelling of DNA-PKcs reported previously (PDB 5LUQ12). Green illustrates regions where the two structures align exactly, orange when the sequence is misaligned by 1-3 residues and red when there is a change of 4+ residues.
Extended Data Fig. 4 Structure of state 2 DNA-PKcs showing key areas of modelled density.
The most extensively remodeled region is part of the circular cradle and is coloured purple. 1 and 2 show two areas previously unmodelled between residues 2573-2594 and 2769-2785, larger side chains are labelled, and residues shown as green sticks. 3 shows a loop previously not modelled between residues 890-907. 4 and 5 illustrate two areas where there is a continuous loop that was previously assumed to be a much larger disordered region. 6. Shows residues 1798-1818 as a continuous loop which was previously modelled as an unconnected helix.
Extended Data Fig. 5 Particle distributions, FSC curves and local resolution maps.
Local resolution maps are coloured according to resolutions and colours displayed in the specific key chart for each map. a) DNA-PKcs state 1, b) DNA-PKcs state 2 and c) DNA-PKcs state 3.
Extended Data Fig. 6 Particle distributions, FSC curves and local resolution maps for DNA-PK monomer and dimer.
Local resolution maps are coloured according to resolutions and colours displayed in the specific key chart for each map. a, DNA-PK monomer and b, DNA-PK dimer.
Extended Data Fig. 7 DNA-PKcs structure with extra density and the location of the C-terminal α-helix of Ku80.
a, DNA-PKcs structure with additional density close to the N-terminal arm predicted to correspond to the CTD of Ku80 labelled and highlighted in yellow15 at 5.8 Å resolution (EMD-8752). b, DNA-PKcs structure with a fragment of DNA solved to 3.8 Å resolution following density modification14, displayed in two orientations. c, The structure of PDB entry 5LUQ modelled into our cryo-EM map of DNA-PKcs with Ku80ct194. This structure highlights where the predicted C-terminal α-helix of Ku80 was previously modelled and where two further helices were predicted to bind and belong to Ku80. d, Structure of apo-DNA-PKcs (state 2, showing no extra density for the C-terminal α-helix of Ku80 or the extra helices predicted in 5LUQ (c). e, Cryo-EM structure of DNA-PKcs with Ku80ct194 to 3.8 Å resolution following density modification14 highlighting the C-terminal α-helix of Ku80 and the lack of the extra helices predicted by 5LUQ (identical to d).
Extended Data Fig. 8 Particle distributions, FSC curves and local resolution maps of DNA-PKcs and complexes with DNA and Ku80CTR.
Local resolution maps are coloured according to resolutions and colours displayed in the specific key chart for each map. a, DNA-PKcs with a fragment of DNA b, DNA-PKcs + Ku80CTR.
Supplementary information
Supplementary Video 1
Side view of the movement between DNA-PKcs states 1–3.
Supplementary Video 2
Back view of the movement between DNA-PKcs states 1–3.
Supplementary Video 3
Back view of the movement between DNA-PKcs states 2 and DNA-PK.
Supplementary Video 4
Side view of the movement between DNA-PKcs states 2 and DNA-PK.
Source data
Source Data Fig. 1
Uncropped SDS-PAGE gel image for Extended Data Fig. 1a.
Rights and permissions
About this article
Cite this article
Chaplin, A.K., Hardwick, S.W., Liang, S. et al. Dimers of DNA-PK create a stage for DNA double-strand break repair. Nat Struct Mol Biol 28, 13–19 (2021). https://doi.org/10.1038/s41594-020-00517-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-00517-x
This article is cited by
-
Structural role for DNA Ligase IV in promoting the fidelity of non-homologous end joining
Nature Communications (2024)
-
Human DNA-dependent protein kinase activation mechanism
Nature Structural & Molecular Biology (2023)
-
Proteogenomics of diffuse gliomas reveal molecular subtypes associated with specific therapeutic targets and immune-evasion mechanisms
Nature Communications (2023)
-
Signal-induced enhancer activation requires Ku70 to read topoisomerase1–DNA covalent complexes
Nature Structural & Molecular Biology (2023)
-
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) drives chronic kidney disease progression in male mice
Nature Communications (2023)