Abstract
Ionotropic orphan delta (GluD) receptors are not gated by glutamate or any other endogenous ligand but are grouped with ionotropic glutamate receptors (iGluRs) based on sequence similarity. GluD1 receptors play critical roles in synaptogenesis and synapse maintenance and have been implicated in neuronal disorders, including schizophrenia, cognitive deficits, and cerebral ataxia. Here we report cryo-EM structures of the rat GluD1 receptor complexed with calcium and the ligand 7-chlorokynurenic acid (7-CKA), elucidating molecular architecture and principles of receptor assembly. The structures reveal a non-swapped architecture at the interface of the extracellular amino-terminal domain (ATD) and the ligand-binding domain (LBD). This finding is in contrast with structures of other families of iGluRs, where the dimer partners between the ATD and LBD layers are swapped. Our results demonstrate that principles of architecture and symmetry are not conserved between delta receptors and other iGluRs and provide a molecular blueprint for understanding the functions of the ‘orphan’ class of iGluRs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM density reconstruction and final models were deposited in the Electron Microscopy DataBase (accession code EMD-0744 for the compact conformation and EMD-0773 for the splayed conformation) and in the Protein Data Bank (accession code PDB 6KSS for the compact conformation and PRB 6KSP for the splayed conformation). The raw movie data have been submitted to the EMPIAR database. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files or from the corresponding author upon reasonable request. Source data for Fig. 3a–c and Extended Data Fig. 9e are available with the paper online.
References
Gao, J. et al. Orphan glutamate receptor δ1 subunit required for high-frequency hearing. Mol. Cell. Biol. 27, 4500–4512 (2007).
Yuzaki, M. The δ2 glutamate receptor: a key molecule controlling synaptic plasticity and structure in Purkinje cells. Cerebellum 3, 89–93 (2004).
Yuzaki, M. & Aricescu, A. R. A GluD coming-of-age story. Trends Neurosci. 40, 138–150 (2017).
Twomey, E. C. & Sobolevsky, A. I. Structural mechanisms of gating in ionotropic glutamate receptors. Biochemistry 57, 267–276 (2018).
Greger, I. H. & Mayer, M. L. Structural biology of glutamate receptor ion channels: towards an understanding of mechanism. Curr. Opin. Struct. Biol. 57, 185–195 (2019).
Yamazaki, M., Araki, K., Shibata, A. & Mishina, M. Molecular cloning of a cDNA encoding a novel member of the mouse glutamate receptor channel family. Biochem. Biophys. Res. Commun. 183, 886–892 (1992).
Lomeli, H. et al. The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett. 315, 318–322 (1993).
Schmid, S. M. & Hollmann, M. To gate or not to gate: are the delta subunits in the glutamate receptor family functional ion channels? Mol. Neurobiol. 37, 126–141 (2008).
Hirai, H. et al. Rescue of abnormal phenotypes of the δ2 glutamate receptor-null mice by mutant δ2 transgenes. EMBO Rep. 6, 90–95 (2005).
Yuzaki, M. New insights into the structure and function of glutamate receptors: the orphan receptor δ2 reveals its family’s secrets. Keio J. Med. 52, 92–99 (2003).
Kakegawa, W., Kohda, K. & Yuzaki, M. The δ2 ‘ionotropic’ glutamate receptor functions as a non-ionotropic receptor to control cerebellar synaptic plasticity. J. Physiol. 584, 89–96 (2007).
Kohda, K. et al. The δ2 glutamate receptor gates long-term depression by coordinating interactions between two AMPA receptor phosphorylation sites. Proc. Natl Acad. Sci. USA 110, E948–E957 (2013).
Yadav, R. et al. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS One 7, e32969 (2012).
Kondo, T., Kakegawa, W. & Yuzaki, M. Induction of long-term depression and phosphorylation of the δ2 glutamate receptor by protein kinase C in cerebellar slices. Eur. J. Neurosci. 22, 1817–1820 (2005).
Utine, G. E. et al. A homozygous deletion in GRID2 causes a human phenotype with cerebellar ataxia and atrophy. J. Child Neurol. 28, 926–932 (2013).
Miyoshi, Y. et al. A new mouse allele of glutamate receptor delta 2 with cerebellar atrophy and progressive ataxia. PLoS One 9, e107867 (2014).
Van Schil, K. et al. Early-onset autosomal recessive cerebellar ataxia associated with retinal dystrophy: new human hotfoot phenotype caused by homozygous GRID2 deletion. Genet. Med. 17, 291–299 (2015).
Guo, S.-Z. et al. A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr. Res. 93, 385–390 (2007).
Benamer, N. et al. GluD1, linked to schizophrenia, controls the burst firing of dopamine neurons. Mol. Psychiatry 23, 691–700 (2018).
Liu, J., Gandhi, P. J., Pavuluri, R., Shelkar, G. P. & Dravid, S. M. Glutamate delta-1 receptor regulates cocaine-induced plasticity in the nucleus accumbens. Transl Psychiatry 8, 219 (2018).
Uemura, T. et al. Trans-synaptic interaction of GluRδ2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079 (2010).
Lee, S.-J., Uemura, T., Yoshida, T. & Mishina, M. GluRδ2 assembles four neurexins into trans-synaptic triad to trigger synapse formation. J. Neurosci. 32, 4688–4701 (2012).
Tao, W., Díaz-Alonso, J., Sheng, N. & Nicoll, R. A. Postsynaptic δ1 glutamate receptor assembles and maintains hippocampal synapses via Cbln2 and neurexin. Proc. Natl Acad. Sci. USA 115, E5373–E5381 (2018).
Yasumura, M. et al. Glutamate receptor δ1 induces preferentially inhibitory presynaptic differentiation of cortical neurons by interacting with neurexins through cerebellin precursor protein subtypes. J. Neurochem. 121, 705–716 (2012).
Elegheert, J. et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–299 (2016).
Uemura, T., Mori, H. & Mishina, M. Direct interaction of GluRδ2 with Shank scaffold proteins in cerebellar Purkinje cells. Mol. Cell. Neurosci. 26, 330–341 (2004).
Takeuchi, T. et al. Control of synaptic connection by glutamate receptor δ2 in the adult cerebellum. J. Neurosci. 25, 2146–2156 (2005).
Wollmuth, L. P. et al. The Lurcher mutation identifies δ2 as an AMPA/kainate receptor-like channel that is potentiated by Ca2+. J. Neurosci. 20, 5973–5980 (2000).
Ikeno, K., Yamakura, T., Yamazaki, M. & Sakimura, K. The Lurcher mutation reveals Ca2+ permeability and PKC modification of the GluRδ channels. Neurosci. Res. 41, 193–200 (2001).
Yadav, R., Rimerman, R., Scofield, M. A. & Dravid, S. M. Mutations in the transmembrane domain M3 generate spontaneously open orphan glutamate δ1 receptor. Brain Res. 1382, 1–8 (2011).
Schmid, S. M., Kott, S., Sager, C., Huelsken, T. & Hollmann, M. The glutamate receptor subunit delta2 is capable of gating its intrinsic ion channel as revealed by ligand binding domain transplantation. Proc. Natl Acad. Sci. USA 106, 10320–10325 (2009).
Orth, A., Tapken, D. & Hollmann, M. The delta subfamily of glutamate receptors: characterization of receptor chimeras and mutants. Eur. J. Neurosci. 37, 1620–1630 (2013).
Naur, P. et al. Ionotropic glutamate-like receptor δ2 binds D-serine and glycine. Proc. Natl Acad. Sci. USA 104, 14116–14121 (2007).
Hansen, K. B. et al. Modulation of the dimer interface at ionotropic glutamate-like receptor δ2 by D-serine and extracellular calcium. J. Neurosci. 29, 907–917 (2009).
Kristensen, A. S. et al. Pharmacology and structural analysis of ligand binding to the orthosteric site of glutamate-like GluD2 receptors. Mol. Pharmacol. 89, 253–262 (2016).
Perroy, J. et al. Direct interaction enables cross-talk between ionotropic and group I metabotropic glutamate receptors. J. Biol. Chem. 283, 6799–6805 (2008).
Suryavanshi, P. S. et al. Glutamate delta-1 receptor regulates metabotropic glutamate receptor 5 signaling in the hippocampus. Mol. Pharmacol. 90, 96–105 (2016).
Ady, V. et al. Type 1 metabotropic glutamate receptors (mGlu1) trigger the gating of GluD2 delta glutamate receptors. EMBO Rep. 15, 103–109 (2014).
Dadak, S. et al. mGlu1 receptor canonical signaling pathway contributes to the opening of the orphan GluD2 receptor. Neuropharmacology 115, 92–99 (2017).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Tao, W. et al. Mechanisms underlying the synaptic trafficking of the glutamate delta receptor GluD1. Mol. Psychiatry 24, 1451–1460 (2019).
Nakagawa, T., Cheng, Y., Ramm, E., Sheng, M. & Walz, T. Structure and different conformational states of native AMPA receptor complexes. Nature 433, 545–549 (2005).
Dürr, K. L. et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell 158, 778–792 (2014).
Meyerson, J. R. et al. Structural mechanism of glutamate receptor activation and desensitization. Nature 514, 328–334 (2014).
Jalali-Yazdi, F., Chowdhury, S., Yoshioka, C. & Gouaux, E. Mechanisms for zinc and proton inhibition of the GluN1/GluN2A NMDA receptor. Cell 175, 1520–1532.e15 (2018).
Zhao, Y., Chen, S., Swensen, A. C., Qian, W.-J. & Gouaux, E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364, 355–362 (2019).
Matsuda, K. et al. Transsynaptic modulation of kainate receptor functions by C1q-like proteins. Neuron 90, 752–767 (2016).
Chen, S. et al. Activation and desensitization mechanism of AMPA receptor-TARP complex by cryo-EM. Cell 170, 1234–1246.e14 (2017).
Twomey, E. C., Yelshanskaya, M. V., Grassucci, R. A., Frank, J. & Sobolevsky, A. I. Structural bases of desensitization in AMPA receptor-auxiliary subunit complexes. Neuron 94, 569–580.e5 (2017).
Sobolevsky, A. I., Rosconi, M. P. & Gouaux, E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 (2009).
Tapken, D. et al. The low binding affinity of D-serine at the ionotropic glutamate receptor GluD2 can be attributed to the hinge region. Sci. Reports 7, 46145 (2017).
Shanks, N. F., Maruo, T., Farina, A. N., Ellisman, M. H. & Nakagawa, T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J. Neurosci. 30, 2728–2740 (2010).
Zhao, H. et al. Preferential assembly of heteromeric kainate and AMPA receptor amino terminal domains. eLife 6, e32056 (2017).
Zhao, H. et al. Analysis of high-affinity assembly for AMPA receptor amino-terminal domains. J. Gen. Physiol. 141, 747–749 (2013).
Chaudhry, C., Plested, A. J., Schuck, P. & Mayer, M. L. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proc. Natl Acad. Sci. USA 106, 12329–12334 (2009).
Cheng, S., Seven, A. B., Wang, J., Skiniotis, G. & Özkan, E. Conformational plasticity in the transsynaptic neurexin-cerebellin-glutamate receptor adhesion complex. Structure 24, 2163–2173 (2016).
Song, X. et al. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 556, 515–519 (2018).
Matsuda, K. et al. Cbln1 is a ligand for an orphan glutamate receptor δ2, a bidirectional synapse organizer. Science 328, 363–368 (2010).
Kandiah, E. et al. CM01: a facility for cryo-electron microscopy at the European Synchrotron. Acta Crystallogr. D Struct. Biol. 75, 528–535 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kidmose, R. T. et al. Namdinator—automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004).
Acknowledgements
This work was supported by the Wellcome Trust DBT India Alliance fellowship (grant number IA/I/13/2/501023) awarded to J.K. A.P.B. thanks ICMR (3/1/3/JRF-2014/HRD-86(60237), India for a senior research fellowship. R.V. thanks SERB for an N-PDF fellowship (N-PDF/2016/002621). M. L. Mayer, NIH, Bethesda kindly gifted the various iGluR constructs that were subcloned and used for construct optimization and mutational studies. E. Gouaux (OHSU, Portland) kindly provided the pEG BacMam vector. We acknowledge the European Synchrotron Radiation Facility for provision of microscope time on CM01, and we thank M. Hons for assistance in EM data collection. We thankfully acknowledge the kind help of M. Karuppasamy, EMBL in grid vitrification. Help with initial screening of conditions for grid vitrification from P. J. Peters, R. Ravelli at M4i, Maastricht University, Masstricht, the Netherlands and V. K. Ragunath, National Electron Cryo-Microscopy facility at the Bangalore Life Sciences Cluster (DBT/PR12422/MED/31/287/2014), NCBS, Bangalore is gratefully acknowledged. We are also thankful to A. Kembhavi and K. Vaghmare, The Inter-University Centre for Astronomy and Astrophysics, Pune for helping with transfer and storage of raw EM data.
Author information
Authors and Affiliations
Contributions
A.P.B. optimized the construct and purified protein, did all of the molecular biology and biochemical experiments, and processed EM data with assistance from J.K. Electrophysiology experiments were done by R.V. J.K. supervised the overall project design and its execution. All authors contributed to the analysis and preparation of the manuscript and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
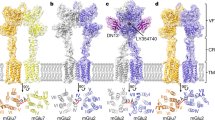
Extended Data Fig. 1 GluD1 purification and cryo-EM data processing.
a, Schematic representation of the optimized GluD1 construct showing the C-terminal truncation at residue 851 and C-terminal thrombin cleavage site, along with GFP and octa-histidine tag (Supplementary Notes). b, Size-exclusion profile of the final purified protein showing receptor stability in optimized buffer conditions. c, Selected 2D class averages from reference-free 2D classification of GluD1 in complex with 1 mM 7-CKA and 1 mM Ca2+. The white arrows mark a few classes that show conformational heterogeneity of the extracellular receptor domains. d, 3D classification of GluD1 into seven classes reveals heterogeneity due to the movement of the two extracellular arms. More details are in Extended Data Fig. 2 and Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM data processing work flow.
A total of 72,149 good particles were obtained by several cycles of 2D class averaging of particles from 4,120 micrographs. The 3D map generated by ab initio 3D reconstruction was further refined heterogeneously into seven conformationally distinct 3D classes. The 3D classes showed heterogeneity due to movement of the two extracellular arms. For the purpose of model building and analysis, a compact (class 5) and a splayed (class 7) conformation map were further refined to a resolution of 8.1 Å and 7.6 Å, respectively.
Extended Data Fig. 3 Local resolution estimates of the cryo-EM maps.
a, d, The sharpened cryo-EM densities of GluD1 Δ851 in 7-CKA and calcium bound form, colored based on local resolution. b, e, Euler angle distribution of particles for the two models is shown. c, f, FSC curves for the cryo-EM maps with mask (red) without mask (blue). The resolution of map corresponding to FSC 0.5 and 0.143 is indicated.
Extended Data Fig. 4 Splayed conformation of GluD1 receptor.
a−f, Architecture of splayed conformation of GluD1 receptors in complex with 7-CKA and calcium. a, Side view highlighting the broadest face of the Y-shaped receptor and 90° rotated views of the sharpened 3D density map. Each subunit is depicted in a different color. The EM reconstructions clearly show the non-swapped arrangement of the ATD and LBD layers. The distances between the centroids (R1-R1 of ATD domains) for AB and CD dimer pairs are shown above the model. The vertical separation between the COMs of ATD dimers and LBD dimers are also shown. Panel b shows the segmented density map for subunits A and B fitted with protein coordinates. c, Superimposition of subunits B/D, B/C, A/D and A/C are shown highlighting similar AB and BC conformations. Helices and sheets are represented as pipes and planks, respectively. Top views of ATD (d), LBD (e) and TM domains (f) are shown. The distances and the angles subtended between the COM of various subunits were measured and are indicated below the top views. More details are in Extended Data Figs. 5 and 6.
Extended Data Fig. 5 Architecture and domain arrangement in compact GluD1 model.
a, Cryo-EM density map of compact GluD1 model is shown in a view parallel to the membrane. The four subunits A, B, C, D are colored in orange, green, yellow and cyan, respectively. The colored spheres represent the COM of the ATD and LBD domains. b, Top view of ATD with segmented EM density map fitted with atomic models is shown. The distances between the COMs of ATDs shown with dashed lines below the EM density map, depicting the arrangement of ATDs in the plane. c, Densities corresponding to LBDs fitted with atomic models. d, The distances from COMs of ATD and LBD are shown. The LBD plane is depicted as a circular disk, and the ATD plane is shown as a dashed ellipse. e,f, Side and top views of angles subtended by COMs of ATD with the COM of the LBD layer. COM plane of the LBD layer is indicated by metallic disk.
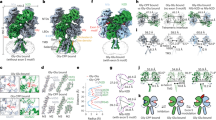
Extended Data Fig. 6 Architecture and domain arrangement in splayed GluD1 model.
a, Cryo-EM density map of splayed GluD1 model is shown in a view parallel to the membrane. The four subunits A, B, C, D are colored in orange, green, yellow and cyan, respectively. The colored spheres represent the COM of ATD and LBD domains. b, Top view of ATD with segmented EM density map fitted with atomic models is shown. The distances between the COMs of ATDs are shown with dashed lines below the EM density map, depicting the arrangement of ATDs in the plane. c, Densities corresponding to LBDs fitted with atomic models is shown. d, The distances from COMs of ATD and LBD are shown. The LBD plane is depicted as a circular disk, and the ATD plane is shown as a dashed ellipse. e,f, Side and top views of angles subtended by COMs of ATD with the COM of the LBD layer. COM plane of the LBD layer is indicated by metallic disk.
Extended Data Fig. 7 Domain arrangement in GluD1, GluA2, GluK2, GluN1/GluN2A and GluN1/GluN2B receptors.
a, Top views of the ATD (a), LBD (b) and TM domains (c) are shown for GluD1, GluA2, GluK2, GluN1/GluN2A and GluN1/GluN2B receptors highlighting the subunit arrangement. Each chain is uniquely colored, and domain arrangement is also depicted in cartoon below each layer. Comparisons for compact and super-splayed conformations of NMDA receptors with that of GluD1 are shown highlighting the fact that in all the conformations of AMPA, kainate and NMDA receptors, the domain swapping between the ATD and LBD layers exists unlike that in GluD1.
Extended Data Fig. 8 Buried surface area between the subdomains.
Surface illustration of the isolated subdomains in gray with buried surface represented in green. The calculated buried surface area for the various domains is also shown. a–d, Analysis for ATD dimer, LBD dimer, ATD dimer-of-dimer and LBD dimer-of-dimer interface for the compact GluD1 model.
Extended Data Fig. 9 C-terminal truncation does not affect assembly of GluD1 receptors.
a, Representative traces for the whole-cell recording of the GluD1, GluD1 Δ851, GluD1(K2LBD) and GluD1(K2LBD) Δ851 expressed in HEK-293T cells are shown in response to 10 mM glutamate application. b–e, Whole-cell patch-clamp recordings (holding potential = −60 mV) from constitutively active GluD1 A634C point mutant receptors. The seal resistance before entering into the whole-cell configuration was always at least 1 GΩ. b, No spontaneous currents were observed for wild-type GluD1 or GluD1 Δ851 receptors, and no effect was observed on 2 mM Ca2+ application. c,d, Overlay of representative traces showing application of either NMDG solution or 1 mM 7-CKA (red), 10 mM d-serine (green) or 2 mM CaCl2 (blue). Dashed line indicates zero current level achieved by application of impermeant NMDG, which blocks the constitutive inward currents for both GluD1 A634C (c) and GluD1 Δ851 A634C receptors (d). The constitutive currents are also modestly inhibited by D-ser or 7-CKA application and potentiated by Ca2+ (c,d) for both the full-length and CT truncated GluD1 receptors. e, Percent inhibition of spontaneous currents by 7-CKA and d-serine calculated with respect to NMDG inhibition. Data for graphs are available as Source data. The number of cells used for the recordings is shown. The error bars represent standard error from the mean.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and Supplementary Notes
Source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 3
Western blots
Source Data Extended Data Fig. 9
Statistical source data
Rights and permissions
About this article
Cite this article
Burada, A.P., Vinnakota, R. & Kumar, J. Cryo-EM structures of the ionotropic glutamate receptor GluD1 reveal a non-swapped architecture. Nat Struct Mol Biol 27, 84–91 (2020). https://doi.org/10.1038/s41594-019-0359-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0359-y
This article is cited by
-
GRID1/GluD1 homozygous variants linked to intellectual disability and spastic paraplegia impair mGlu1/5 receptor signaling and excitatory synapses
Molecular Psychiatry (2024)
-
Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor
Nature (2023)
-
Multi-omics approaches identify novel prognostic biomarkers of autophagy in uveal melanoma
Journal of Cancer Research and Clinical Oncology (2023)