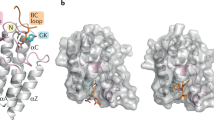
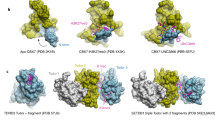
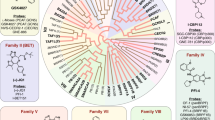
Abstract
The bromodomain (BrD) is a conserved structural module found in chromatin- and transcription-associated proteins that acts as the primary reader for acetylated lysine residues. This basic activity endows BrD proteins with versatile functions in the regulation of protein-protein interactions mediating chromatin-templated gene transcription, DNA recombination, replication and repair. Consequently, BrD proteins are involved in the pathogenesis of numerous human diseases. In this Review, we highlight our current understanding of BrD biology, and discuss the latest development of small-molecule inhibitors targeting BrDs as emerging epigenetic therapies for cancer and inflammatory disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Allfrey, V. G., Faulkner, R. & Mirsky, A. E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51, 786–794 (1964).
Brownell, J. E. et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 (1996).
Kuo, M. H. et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 (1996).
Taunton, J., Hassig, C. A. & Schreiber, S. L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999). First three-dimensional structure of the BrD and discovery of the BrD as the lysine-acetylated histone reader.
Haynes, S. R. et al. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20, 2603 (1992).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000). First presentation of the histone code hypothesis.
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). Demonstration of BET family BRD4 protein as a new anticancer drug target.
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010). Demonstration of BET family BRD4 protein as a new anti-inflammation drug target.
Smith, S. G. & Zhou, M. M. The bromodomain: a new target in emerging epigenetic medicine. ACS Chem. Biol. 11, 598–608 (2016).
Filippakopoulos, P. & Knapp, S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 13, 337–356 (2014).
Tamkun, J. W. et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68, 561–572 (1992).
Presnell, S. R. & Cohen, F. E. Topological distribution of four-alpha-helix bundles. Proc. Natl. Acad. Sci. USA 86, 6592–6596 (1989).
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Owen, D. J. et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19, 6141–6149 (2000). Establishing the key determinants of Kac recognition by the BrD.
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012). Demonstrating the conserved left-handed four-helix bundle structure for human BrDs.
Zeng, L. & Zhou, M. M. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513, 124–128 (2002).
Dancy, B. M. & Cole, P. A. Protein lysine acetylation by p300/CBP. Chem. Rev. 115, 2419–2452 (2015).
Ortega, E. et al. Transcription factor dimerization activates the p300 acetyltransferase. Nature 562, 538–544 (2018).
Burley, S. K. & Roeder, R. G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 (1996).
Avvakumov, N. & Côté, J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26, 5395–5407 (2007).
Vezzoli, A. et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 17, 617–619 (2010).
Ullah, M. et al. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 28, 6828–6843 (2008).
Perez-Campo, F. M., Borrow, J., Kouskoff, V. & Lacaud, G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood 113, 4866–4874 (2009).
Panagopoulos, I. et al. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22; p13). Hum. Mol. Genet. 10, 395–404 (2001).
Mishima, Y. et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 118, 2443–2453 (2011).
Monden, T. et al. p120 acts as a specific coactivator for 9-cis-retinoic acid receptor (RXR) on peroxisome proliferator-activated receptor-gamma/RXR heterodimers. Mol. Endocrinol. 13, 1695–1703 (1999).
Yamada, H. Y. & Rao, C. V. BRD8 is a potential chemosensitizing target for spindle poisons in colorectal cancer therapy. Int. J. Oncol. 35, 1101–1109 (2009).
Mashtalir, N. et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 175, 1272–1288.e20 (2018).
Ho, L. & Crabtree, G. R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Dirscherl, S. S. & Krebs, J. E. Functional diversity of ISWI complexes. Biochem. Cell Biol. 82, 482–489 (2004).
Thompson, P. J., Norton, K. A., Niri, F. H., Dawe, C. E. & McDermid, H. E. CECR2 is involved in spermatogenesis and forms a complex with SNF2H in the testis. J. Mol. Biol. 415, 793–806 (2012).
Xiao, A. et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62 (2009).
Koo, S. J. et al. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget 7, 70323–70335 (2016).
Wu, S. Y. & Chiang, C. M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 282, 13141–13145 (2007).
Rahman, S. et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 31, 2641–2652 (2011).
Shen, C. et al. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell 60, 847–859 (2015).
Zhang, Q. et al. Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure 24, 1201–1208 (2016).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Morinière, J. et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461, 664–668 (2009).
Shi, J. et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25, 210–225 (2014).
Schröder, S. et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem. 287, 1090–1099 (2012).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Jain, A. K. & Barton, M. C. Regulation of p53: TRIM24 enters the RING. Cell Cycle 8, 3668–3674 (2009).
Sanchez, R. & Zhou, M. M. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36, 364–372 (2011).
Zeng, L. et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 15, 626–633 (2008).
Ivanov, A. V. et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 (2007).
Xi, Q. et al. A poised chromatin platform for TGF-β access to master regulators. Cell 147, 1511–1524 (2011).
Xue, J. et al. Tumour suppressor TRIM33 targets nuclear β-catenin degradation. Nat. Commun. 6, 6156 (2015).
Khetchoumian, K. et al. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J. Biol. Chem. 279, 48329–48341 (2004).
Gibson, T. J., Ramu, C., Gemünd, C. & Aasland, R. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem. Sci. 23, 242–244 (1998).
Granito, A. et al. PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am. J. Gastroenterol. 105, 125–131 (2010).
Seeler, J. S. et al. Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol. Cell. Biol. 21, 3314–3324 (2001).
Bottomley, M. J. et al. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8, 626–633 (2001).
Fossey, S. C. et al. Identification and characterization of PRKCBP1, a candidate RACK-like protein. Mamm. Genome 11, 919–925 (2000).
Malovannaya, A. et al. Analysis of the human endogenous coregulator complexome. Cell 145, 787–799 (2011).
Ansieau, S. & Sergeant, A. BS69 and RACK7, a potential novel class of tumor suppressor genes. Pathol. Biol. (Paris) 51, 397–399 (2003).
Wen, H. et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014).
Savitsky, P. et al. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 17, 2724–2737 (2016).
Yao, N. et al. The structure of the ZMYND8/Drebrin complex suggests a cytoplasmic sequestering mechanism of ZMYND8 by Drebrin. Structure 25, 1657–1666.e3 (2017).
Li, D. & Roberts, R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58, 2085–2097 (2001).
Huang, H., Rambaldi, I., Daniels, E. & Featherstone, M. Expression of the Wdr9 gene and protein products during mouse development. Dev. Dyn. 227, 608–614 (2003).
Kim, Y. J. et al. WDR11-mediated Hedgehog signalling defects underlie a new ciliopathy related to Kallmann syndrome. EMBO Rep. 19, 269–289 (2018).
Zeng, L. et al. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J. Am. Chem. Soc. 127, 2376–2377 (2005). First report of small-molecule BrD inhibitors and the druggability of the BrD.
Sachchidanand et al. Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem. Biol. 13, 81–90 (2006).
Adachi, K. et al. Thienotriazolodiazepine compound and medicinal use thereof. PCT/JP2006/310709 (Mitsubishi Tanabe Pharma Corporation, Japan, 2006).
Miyoshi, S., Ooike, S., Iwata, K., Hikawa, H. & Sugahara, K. Antitumor agent. US 2010/0286127 A1 (Mitsubishi Tanabe Pharma Corporation, Japan, 2008).
Smith, S. G., Sanchez, R. & Zhou, M.-M. Privileged diazepine compounds and their emergence as bromodomain inhibitors. Chem. Biol. 21, 573–583 (2014).
French, C. A. et al. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63, 304–307 (2003).
Zhang, G. et al. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J. Biol. Chem. 287, 28840–28851 (2012).
Moros, A. et al. Synergistic antitumor activity of lenalidomide with the BET bromodomain inhibitor CPI203 in bortezomib-resistant mantle cell lymphoma. Leukemia 28, 2049–2059 (2014).
Wong, C. et al. The bromodomain and extra-terminal inhibitor CPI203 enhances the antiproliferative effects of rapamycin on human neuroendocrine tumors. Cell Death Dis. 5, e1450 (2014).
Noel, J. K. et al. Development of the BET bromodomain inhibitor OTX015. Mol. Cancer Ther. 12, C244 (2013).
Hewings, D. S. et al. 3,5-dimethylisoxazoles act as acetyl-lysine-mimetic bromodomain ligands. J. Med. Chem. 54, 6761–6770 (2011).
Wyspiańska, B. S. et al. BET protein inhibition shows efficacy against JAK2V617F-driven neoplasms. Leukemia 28, 88–97 (2014).
Gosmini, R. et al. The discovery of I-BET726 (GSK1324726A), a potent tetrahydroquinoline ApoA1 up-regulator and selective BET bromodomain inhibitor. J. Med. Chem. 57, 8111–8131 (2014).
Picaud, S. et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 73, 3336–3346 (2013).
Picaud, S. et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc. Natl. Acad. Sci. USA 110, 19754–19759 (2013).
Cheung, K. L. et al. Distinct roles of Brd2 and Brd4 in potentiating the transcriptional program for Th17 cell differentiation. Mol. Cell 65, 1068–1080.e5 (2017).
Schröder, S. et al. Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol. Cell 52, 314–324 (2013).
Cheung, K. et al. BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc. Natl. Acad. Sci. USA 114, 2952–2957 (2017).
Gacias, M. et al. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem. Biol. 21, 841–854 (2014).
Vidler, L. R., Brown, N., Knapp, S. & Hoelder, S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J. Med. Chem. 55, 7346–7359 (2012).
Wu, Q. et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat. Commun. 10, 1915 (2019).
Demont, E. H. et al. 1,3-Dimethyl benzimidazolones are potent, selective inhibitors of the BRPF1 bromodomain. ACS Med. Chem. Lett. 5, 1190–1195 (2014).
Palmer, W. S. et al. Structure-guided design of IACS-9571, a selective high-affinity dual TRIM24-BRPF1 bromodomain inhibitor. J. Med. Chem. 59, 1440–1454 (2016).
Demont, E. H. et al. Fragment-based discovery of low-micromolar ATAD2 bromodomain inhibitors. J. Med. Chem. 58, 5649–5673 (2015).
Harner, M. J., Chauder, B. A., Phan, J. & Fesik, S. W. Fragment-based screening of the bromodomain of ATAD2. J. Med. Chem. 57, 9687–9692 (2014).
Bamborough, P. et al. A Chemical Probe for the ATAD2 Bromodomain. Angew. Chem. Int. Ed. 55, 11382–11386 (2016).
Drouin, L. et al. Structure enabled design of BAZ2-ICR, a chemical probe targeting the bromodomains of BAZ2A and BAZ2B. J. Med. Chem. 58, 2553–2559 (2015).
Ferguson, F. M. et al. Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain. J. Med. Chem. 56, 10183–10187 (2013).
Chen, P. et al. Discovery and characterization of GSK2801, a selective chemical probe for the bromodomains BAZ2A and BAZ2B. J. Med. Chem. 59, 1410–1424 (2016).
Clark, P. G. K. et al. LP99: discovery and synthesis of the first selective BRD7/9 bromodomain inhibitor. Angew. Chem. Int. Ed. 54, 6217–6221 (2015).
Hay, D. A. et al. Design and synthesis of potent and selective inhibitors of BRD7 and BRD9 bromodomains. MedChemComm 6, 1381–1386 (2015).
Picaud, S. et al. 9H-purine scaffold reveals induced-fit pocket plasticity of the BRD9 bromodomain. J. Med. Chem. 58, 2718–2736 (2015).
Theodoulou, N. H. et al. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 59, 14225–1439 (2016).
Arnold, L. D., Foreman, K. W., Jin, M., Wanner, J. & Werner, D. Bromodomain ligands capable of dimerizing in an aqueous solution, and methods of using same. US patent 20140243286 (2014).
Waring, M. J. et al. Potent and selective bivalent inhibitors of BET bromodomains. Nat. Chem. Biol. 12, 1097–1104 (2016).
Tanaka, M. et al. Design and characterization of bivalent BET inhibitors. Nat. Chem. Biol. 12, 1089–1096 (2016).
Ren, C. et al. Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth. Proc. Natl. Acad. Sci. USA 115, 7949–7954 (2018).
Amemiya, S., Yamaguchi, T., Hashimoto, Y. & Noguchi-Yachide, T. Synthesis and evaluation of novel dual BRD4/HDAC inhibitors. Bioorg. Med. Chem. 25, 3677–3684 (2017).
Ember, S. W. J. et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem. Biol. 9, 1160–1171 (2014).
Cromm, P. M. & Crews, C. M. Targeted protein degradation: from chemical biology to drug discovery. Cell Chem. Biol. 24, 1181–1190 (2017).
Nowak, R. P. et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 14, 706–714 (2018).
Lu, J. et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22, 755–763 (2015).
Remillard, D. et al. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew. Chem. Int. Ed. 56, 5738–5743 (2017).
Gechijian, L. N. et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat. Chem. Biol. 14, 405–412 (2018).
Mertz, J. A. et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 108, 16669–16674 (2011).
Lockwood, W. W., Zejnullahu, K., Bradner, J. E. & Varmus, H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc. Natl. Acad. Sci. USA 109, 19408–19413 (2012).
Puissant, A. et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 (2013).
Cheng, Z. et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 19, 1748–1759 (2013).
Asangani, I. A. et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 (2014).
Boehm, D. et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle 12, 452–462 (2013).
Mele, D. A. et al. BET bromodomain inhibition suppresses TH17-mediated pathology. J. Exp. Med. 210, 2181–2190 (2013).
Anand, P. et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 (2013).
Resverlogix. News Release - Resverlogix officially attains phase 3 status with a European regulatory authority. https://www.resverlogix.com/investors/news?article=521 (2015).
Lewin, J. et al. Phase Ib trial with birabresib, a small-molecule inhibitor of bromodomain and extraterminal proteins, in patients with selected advanced solid tumors. J. Clin. Oncol. 36, 3007–3014 (2018). Key clinical evaluation of pharmacological inhibition of BET BrD proteins for cancer treatment as well as associated dose-limiting toxicities.
Zeng, L. et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–262 (2010). Another Kac histone reader domain distinct from the BrD.
Li, Y. et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159, 558–571 (2014). First report of the YEATS domain as acyl-lysine histone reader in gene transcription.
Andrews, F. H., Shanle, E. K., Strahl, B. D. & Kutateladze, T. G. The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation. Transcription 7, 14–20 (2016).
Arrowsmith, C. H. & Schapira, M. Targeting non-bromodomain chromatin readers. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-019-0290-2 (2019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.-M.Z. is a founder, director and shareholder of Parkside Scientific Inc.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–3
Rights and permissions
About this article
Cite this article
Zaware, N., Zhou, MM. Bromodomain biology and drug discovery. Nat Struct Mol Biol 26, 870–879 (2019). https://doi.org/10.1038/s41594-019-0309-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0309-8
This article is cited by
-
Targeting histone modifiers in bladder cancer therapy — preclinical and clinical evidence
Nature Reviews Urology (2024)
-
Targeted protein degradation via intramolecular bivalent glues
Nature (2024)
-
Targeting bromodomain-containing proteins: research advances of drug discovery
Molecular Biomedicine (2023)
-
Defining super-enhancers by highly ranked histone H4 multi-acetylation levels identifies transcription factors associated with glioblastoma stem-like properties
BMC Genomics (2023)
-
Bromodomain and extraterminal (BET) proteins: biological functions, diseases, and targeted therapy
Signal Transduction and Targeted Therapy (2023)