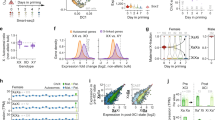
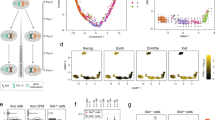
Abstract
Ohno's hypothesis postulates that upregulation of X-linked genes rectifies their dosage imbalance relative to autosomal genes, which are present in two active copies per cell. Here we have dissected X-chromosome upregulation into the kinetics of transcription, inferred from allele-specific single-cell RNA sequencing data from somatic and embryonic mouse cells. We confirmed increased X-chromosome expression levels in female and male cells and found that the X chromosome achieved upregulation by elevated burst frequencies. By monitoring transcriptional kinetics in differentiating female mouse embryonic stem cells, we found that increased burst frequency was established on the active X chromosome when X inactivation took place on the other allele. Thus, our study provides mechanistic insights into X-chromosome upregulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The sequencing data are available at the European Nucleotide Archive (E-MTAB-7098), ArrayExpress (E-MTAB-6385) and GEO (GSE74155).
Code availability
The computational code used for calculations and plotting of data is available at https://sourceforge.net/projects/kinetics-of-x-upregulation.
References
Berta, P. et al. Genetic-evidence equating SRY and the testis-determining factor. Nature 348, 448–450 (1990).
Wallis, M. C., Waters, P. D. & Graves, J. A. M. Sex determination in mammals—before and after the evolution of SRY. Cell. Mol. Life Sci. 65, 3182–3195 (2008).
Charlesworth, B. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6, 149–162 (1996).
Ohno, S. Sex Chromosomes and Sex-linked Genes (Springer, 1967).
Lyon, M. F. Gene action in X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961).
Gupta, V. et al. Global analysis of X-chromosome dosage compensation. J. Biol. 5, 3 (2006).
Nguyen, D. K. & Disteche, C. M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38, 47–53 (2006).
Lin, H. et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 5, 2809–2820 (2007).
Lin, H. et al. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat. Genet. 43, 1169–1170 (2011); author reply 43, 1171–1172.
Deng, X. et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43, 1179–1185 (2011).
Julien, P. et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10, e1001328 (2012).
Xiong, Y. et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 42, 1043–1047 (2010).
Wang, M., Lin, F., Xing, K. & Liu, L. Random X-chromosome inactivation dynamics in vivo by single-cell RNA sequencing. BMC Genom. 18, 90 (2017).
Lin, F. Q., Xing, K., Zhang, J. Z. & He, X. L. Expression reduction in mammalian X chromosome evolution refutes Ohno's hypothesis of dosage compensation. Proc. Natl Acad. Sci. USA 109, 11752–11757 (2012).
Li, X. et al. Dosage compensation in the process of inactivation/reactivation during both germ cell development and early embryogenesis in mouse. Sci. Rep. 7, 3729 (2017).
Sangrithi, M. N. et al. Non-canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev. Cell 40, 289–301.e3 (2017).
Yang, J.R. et al. Dosage sensitivity of X-linked genes in human embryonic single cells. BMC Genom. 20, 42 (2019).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–U183 (2012).
Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4 (2017).
Levine, M. & Tjian, R. Transcription regulation and animal diversity. Nature 424, 147–151 (2003).
Raj, A. & van Oudenaarden, A. Nature, nurture or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Larsson, A. J. M. et al. Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254 (2019).
Symmons, O. et al. Allele-specific RNA imaging shows that allelic imbalances can arise in tissues through transcriptional bursting. PLoS Genet. 15, e1007874 (2019).
Peccoud, J. & Ycart, B. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 48, 222–234 (1995).
Mank, J. E. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29, 677–683 (2013).
Deng, X. et al. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life and MOF-mediated H4K16 acetylation. Dev. Cell 25, 55–68 (2013).
Faucillion, M. L. & Larsson, J. Increased expression of X-linked genes in mammals is associated with a higher stability of transcripts and an increased ribosome density. Genome Biol. Evol. 7, 1039–1052 (2015).
Augui, S., Nora, E. P. & Heard, E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442 (2011).
Wutz, A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12, 542–553 (2011).
Chen, G. et al. Single-cell analyses of X chromosome inactivation dynamics and pluripotency during differentiation. Genome Res. 26, 1342–1354 (2016).
Pintacuda, G. & Cerase, A. X inactivation lessons from differentiating mouse embryonic stem cells. Stem Cell Rev. Rep. 11, 699–705 (2015).
Deng, X., Berletch, J. B., Nguyen, D. K. & Disteche, C. M. X-chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15, 367–378 (2014).
Yang, F., Babak, T., Shendure, J. & Disteche, C. M. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20, 614–622 (2010).
Yildirim, E., Sadreyev, R. I., Pinter, S. F. & Lee, J. T. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat. Struct. Mol. Biol. 19, 56–U72 (2012).
Bartman, C. R. et al. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell 73, 519–532.e4 (2019).
Bartman, C. R., Hsu, S. C., Hsiung, C. C. S., Raj, A. & Blobel, G. A. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell 62, 237–247 (2016).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Reinius, B. et al. Analysis of allelic expression patterns in clonal somatic cells by single-cell RNA-seq. Nat. Genet. 48, 1430–1435 (2016).
Herzog, V. A. et al. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017).
Li, B. et al. A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Sci. Rep. 7, 4200 (2017).
Chadwick, L. H., Pertz, L. M., Broman, K. W., Bartolomei, M. S. & Willard, H. F. Genetic control of X chromosome inactivation in mice: definition of the Xce candidate interval. Genetics 173, 2103–2110 (2006).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Acknowledgements
This study was made possible by grants to B.R. from the Ragnar Söderberg Foundation, the Swedish Research Council (2017-01723) and Åke Wiberg’s Foundation; and grants to R.S. from the European Research Council (648842) and the Swedish Research Council (2017-01062). The authors thank D. Camsund for providing insightful comments to the manuscript.
Author information
Authors and Affiliations
Contributions
B.R. conceived and supervised the study. A.J.M.L. and B.R. analyzed the data and wrote the manuscript. A.J.M.L., C.C., R.S. and B.R. participated in interpreting the data and editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Larsson, A.J.M., Coucoravas, C., Sandberg, R. et al. X-chromosome upregulation is driven by increased burst frequency. Nat Struct Mol Biol 26, 963–969 (2019). https://doi.org/10.1038/s41594-019-0306-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0306-y
This article is cited by
-
RNA stability controlled by m6A methylation contributes to X-to-autosome dosage compensation in mammals
Nature Structural & Molecular Biology (2023)
-
Elastic dosage compensation by X-chromosome upregulation
Nature Communications (2022)
-
Mammalian X-chromosome inactivation: proposed role in suppression of the male programme in genetic females
Journal of Genetics (2022)
-
Enhanced chromatin accessibility contributes to X chromosome dosage compensation in mammals
Genome Biology (2021)
-
Integrated analysis of Xist upregulation and X-chromosome inactivation with single-cell and single-allele resolution
Nature Communications (2021)