Abstract
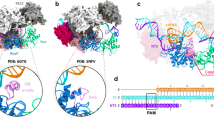
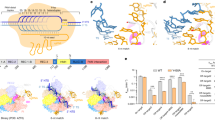
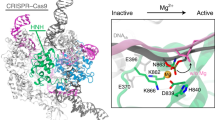
The RNA-guided Cas9 endonuclease from Streptococcus pyogenes is a single-turnover enzyme that displays a stable product state after double-stranded-DNA cleavage. Here, we present cryo-EM structures of precatalytic, postcatalytic and product states of the active Cas9–sgRNA–DNA complex in the presence of Mg2+. In the precatalytic state, Cas9 adopts the ‘checkpoint’ conformation with the HNH nuclease domain positioned far away from the DNA. Transition to the postcatalytic state involves a dramatic ~34-Å swing of the HNH domain and disorder of the REC2 recognition domain. The postcatalytic state captures the cleaved substrate bound to the catalytically competent HNH active site. In the product state, the HNH domain is disordered, REC2 returns to the precatalytic conformation, and additional interactions of REC3 and RuvC with nucleic acids are formed. The coupled domain motions and interactions between the enzyme and the RNA-DNA hybrid provide new insights into the mechanism of genome editing by Cas9.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data needed to assess and evaluate the conclusions in the paper are available in the main text and supplementary information. The coordinates and electron density maps are deposited in the Protein Data Bank and EMDB with the following accession numbers (respectively): 6O0Z and 0585 for precatalytic complex (state I), 6O0Y and 0584 for postcatalytic complex (state II) and 6O0X and 0583 for product complex (state III). Uncropped gel images for Supplementary Fig. 3 are shown in Supplementary Data Set 1. All other data are available upon request.
References
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Boyle, E. A. et al. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl Acad. Sci. USA 114, 5461–5466 (2017).
Sternberg, S. H., LaFrance, B., Kaplan, M. & Doudna, J. A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 (2015).
Dagdas, Y. S., Chen, J. S., Sternberg, S. H., Doudna, J. A. & Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3, eaao0027 (2017).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Sung, K., Park, J., Kim, Y., Lee, N. K. & Kim, S. K. Target specificity of Cas9 nuclease via DNA rearrangement regulated by the REC2 domain. J. Am. Chem. Soc. 140, 7778–7781 (2018).
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014).
Jiang, F. et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014).
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. Striking plasticity of CRISPR-Cas9 and key role of non-target DNA, as revealed by molecular simulations. ACS Cent. Sci. 2, 756–763 (2016).
Gong, S., Yu, H. H., Johnson, K. A. & Taylor, D. W. DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 22, 359–371 (2018).
Raper, A. T., Stephenson, A. A. & Suo, Z. Functional insights revealed by the kinetic mechanism of CRISPR/Cas9. J. Am. Chem. Soc. 140, 2971–2984 (2018).
Palermo, G., Miao, Y., Walker, R. C., Jinek, M. & McCammon, J. A. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl Acad. Sci. USA 114, 7260–7265 (2017).
Zuo, Z. & Liu, J. Structure and dynamics of Cas9 HNH domain catalytic state. Sci. Rep. 7, 17271 (2017).
Brinkman, E. K. et al. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol. Cell 70, 801–813 e806 (2018).
Clarke, R. et al. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol. Cell 71, 42–55.e48 (2018).
Jiang, F., Zhou, K., Ma, L., Gressel, S. & Doudna, J. A. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9, 1911 (2018).
Wilkinson, R. A., Martin, C., Nemudryi, A. A. & Wiedenheft, B. CRISPR RNA-guided autonomous delivery of Cas9. Nat. Struct. Mol. Biol. 26, 14–24 (2019).
Shen, B. W., Landthaler, M., Shub, D. A. & Stoddard, B. L. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 342, 43–56 (2004).
Anders, C. & Jinek, M. In vitro enzymology of Cas9. Meth. Enzymol. 546, 1–20 (2014).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Acknowledgements
This work was supported by the intramural research program of the National Cancer Institute (X.Z., S.C. and S.S.), NIH grants (no. GM097042 to M.S. and no. HD081534 to B.J.M.), UIC Center for Clinical and Translational Sciences (R.C. and A.K.P.) and a Canada Excellence Research Chair Award (to S.S.).
Author information
Authors and Affiliations
Contributions
R.C., B.J.M., M.S. and S.S. conceived the project. R.C. and A.K.P. purified the complex. S.C. prepared cryo-EM grids. A.M. collected cryo-EM data. X.Z. carried out cryo-EM image processing. X.Z., A.K.P. and M.S. built and refined atomic models. M.S. and S.S. provided overall supervision and guidance at all stages of the project. All authors contributed to the experimental design and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Representative cryo-EM density maps and models corresponding to states I-III.
(a) Atomic-resolution models of states I-III of the Cas9-sgRNA-dsDNA ternary complex. Arginine-rich bridge of Cas9 and PAM-distal DNA duplex are shown in the upper and lower panels, respectively. (b) Unsharpened (top) and sharpened (bottom) EM maps for each structure are presented. Coloring scheme is the same as in Fig. 1.
Supplementary Figure 2 Steric occlusion of NTS from the tunnel between HNH and RuvC in state I.
(Left) Close-up view of the HNH-RuvC interface where the two domains create a tunnel for the NTS in the previously reported “Mg2+-free” ternary complex crystal structure (5F9R). (Right) Close-up view of the same tunnel in state I, showing closure of the gap, making it inaccessible for the NTS.
Supplementary Figure 3 In vitro analyses of the Cas9-sgRNA-DNA ternary complex.
(a) Assessment of Cas9 stability during DNA cleavage. The ternary complex sample (22.5 μL) corresponding to each timepoint was loaded onto a 4–20% precast SDS-PA gel. The gel shows lack of proteolytic degradation of Cas9 even after 960 min at +37 °C. (b) Probing the ternary complex stability and DNA cleavage levels using an EMSA. Untreated and RNaseA/proteinase K-treated samples (18 μL) corresponding to each time point were analyzed on an 8% 0.5x TBE-PA gel. The same gel was stained for nucleic acids (left) and then for protein (right). The ternary complex is stable after addition of Mg2+ and exhibits single-turnover kinetics until the 240 min timepoint, thus explaining how different structural states of the complex may have been captured at 30 min. (c) Analysis of the Cas9-catalyzed DNA cleavage. Untreated and RNaseA/proteinase K-treated samples (20 μL) corresponding to each time point were analyzed on a denaturing 15% TBE-urea PA gel. The gel demonstrates that Cas9 is catalytically active. Control samples contained 400 nM Cas9, 600 nM sgRNA, or 600 nM DNA only. For more detail, see on-line Methods. Uncropped gel images are shown in Supplementary Data Set 1.
Supplementary Figure 4 Interactions between RuvC and extended DNA duplex, and a likely path for the single stranded NTS that closes the R-loop.
(a) In states II and III, the RuvC domain (light blue) interacts with the 10 bp-long extension of the DNA duplex (TS is blue, NTS is purple) via the solvent-exposed loop that carries three positively charged residues. A segment of RuvC encompassing residues 1000–1076 is disordered (dashed line). Structural comparison shows that the longer duplex used in our study would clash with the 1000–1076 segment of the “Mg2+-free” ternary complex (PDB ID 5F9R; grey), thus causing the disorder in this segment in states II and III. (b) Low-resolution cryo-EM density (purple transparent surface) extends from the cleaved NTS (purple cartoon) and threads between RuvC and HNH (pink) domains. The density points towards the distal TS-NTS duplex, which is sandwiched between RuvC and REC3, suggesting that this is the path for the single-stranded NTS that closes the R-loop. sgRNA is orange cartoon, TS is blue cartoon, NTS is purple cartoon, HNH domain is pink transparent surface, and the rest of the Cas9 protein is white transparent surface.
Supplementary Figure 5 Structural changes after Cas9-catalyzed DNA cleavage.
Structural comparison of states II and III of the Cas9-sgRNA-dsDNA ternary complex. HNH and REC2 domains are excluded from analysis because of the alternate structural disorder in two states. Analysis and representation were completed as in Fig. 2a. Cas9 domains and nucleic acids are colored as in Fig. 1a.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Supplementary Notes 1 and 2, and Supplementary Data Set 1
Supplementary Video 1
Conformational changes in the Cas9 ternary complex during transition from state I (‘conformation’ checkpoint) to state II (‘postcatalytic’). HNH domain (pink) swings about the axis perpendicular to the plane of view and it docks at the cleavage site in the TS DNA (blue). Segments of the complex that become disordered (for example, REC2 domain and parts of RuvC) and ordered (for example, loops in REC3 and distal duplex) upon transition, flicker at the start and the end of the video, respectively. Coloring scheme is the same as in Fig. 1.
Supplementary Video 2
Structural rearrangements in the Cas9 ternary complex during transition from state II (‘postcatalytic’) to state III (‘product’). HNH domain (pink) dissociates from cleaved TS DNA (blue) and moves towards solvent, leading to its disorder. REC2 domain (green) now returns to its location as in state I. The REC lobe and distal duplex slightly rotate around the horizontal axis causing the complex to open up a bit more when compared to state II. Segments of the complex that become disordered (for example, HNH and HNH-REC2 linker) and ordered (for example, REC2) upon transition, flicker at the start and the end of the video, respectively. Coloring scheme is the same as in Fig. 1.
Rights and permissions
About this article
Cite this article
Zhu, X., Clarke, R., Puppala, A.K. et al. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol 26, 679–685 (2019). https://doi.org/10.1038/s41594-019-0258-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0258-2
This article is cited by
-
Engineering Cas9: next generation of genomic editors
Applied Microbiology and Biotechnology (2024)
-
Prime editing with genuine Cas9 nickases minimizes unwanted indels
Nature Communications (2023)
-
Genome-wide CRISPR off-target prediction and optimization using RNA-DNA interaction fingerprints
Nature Communications (2023)
-
Coupled catalytic states and the role of metal coordination in Cas9
Nature Catalysis (2023)
-
CRISPR-Combo–mediated orthogonal genome editing and transcriptional activation for plant breeding
Nature Protocols (2023)