Abstract
The cyclic enzymatic removal and ligation of the C-terminal tyrosine of α-tubulin generates heterogeneous microtubules and affects their functions. Here we describe the crystal and solution structure of the tubulin carboxypeptidase complex between vasohibin (VASH1) and small vasohibin-binding protein (SVBP), which folds in a long helix, which stabilizes the VASH1 catalytic domain. This structure, combined with molecular docking and mutagenesis experiments, reveals which residues are responsible for recognition and cleavage of the tubulin C-terminal tyrosine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The coordinates for the crystal structure of the VASH1–SVBP complex are deposited in the PDB under the accession identifier PDB 6NVQ. All other data and the coordinates of the HADDOCK model are available upon request.
References
Wehenkel, A. & Janke, C. Nat. Cell Biol. 16, 303–305 (2014).
Murofushi, H. J. Biochem. 87, 979–984 (1980).
Prota, A. E. et al. J. Cell Biol. 200, 259–270 (2013).
Szyk, A., Deaconescu, A. M., Piszczek, G. & Roll-Mecak, A. Nat. Struct. Mol. Biol. 18, 1250–1258 (2011).
Nieuwenhuis, J. et al. Science 358, eaao5676–1456 (2017).
Aillaud, C. et al. Science 358, 1448–1453 (2017).
Sanchez-Pulido, L. & Ponting, C. P. Bioinformatics 32, 1441–1445 (2016).
Waltersperger, S. et al. J. Synchrotron Radiat. 22, 895–900 (2015).
Rambo, R. P. & Tainer, J. A. Biopolymers 95, 559–571 (2011).
Petoukhov, M. V. & Svergun, D. I. Biophys J 89, 1237–1250 (2005).
van Zundert, G. C. P. et al. J. Mol. Biol. 428, 720–725 (2016).
Otto, H.-H. & Schirmeister, T. Chem. Rev. 97, 133–172 (1997).
Zhu, M., Shao, F., Innes, R. W., Dixon, J. E. & Xu, Z. Proc. Natl Acad. Sci. USA 101, 302–307 (2004).
Luna-Vargas, M. P. A. et al. J. Struct. Biol. 175, 113–119 (2011).
Newman, J. et al. Acta Crystallogr. D Biol. Crystallogr. 61, 1426–1431 (2005).
Nurizzo, D. et al. Acta Crystallogr. D Struct. Biol. 72, 966–975 (2016).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Pape, T. & Schneider, T. J. Synchr. Rad. 37, 843–844 (2004).
Uson, I. & Sheldrick, G. Curr. Opin. Struct. Biol. 9, 643–648 (1999).
Ness, S., de Graaff, R., Abrahams, J. & Pannu, N. Struct. Solut. Struct. (Camb.) 12, 1753–1761 (2004).
Murshudov, G. N. et al. Macromol. Cryst. Struct. 67, 355–367 (2011).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Joosten, R. P., Long, F., Murshudov, G. N. & Perrakis, A. IUCrJ 1, 213–220 (2014).
Chen, V. B. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Petoukhov, M. V. et al. J. Appl. Crystallogr. 45, 342–350 (2012).
Svergun, D., Barberato, C. & Koch, M. H. J. J. Appl. Crystallogr. 28, 768–773 (1995).
Dominguez, C., Boelens, R. & Bonvin, A. M. J. J. J. Am. Chem. Soc. 125, 1731–1737 (2003).
Holm, L. & Laakso, L. M. Nucleic Acids Res. 44, W351–W355 (2016).
Sievers, F. et al. Mol. Syst. Biol. 7, 539–539 (2011).
Chojnacki, S., Cowley, A., Lee, J., Foix, A. & Lopez, R. Nucleic Acids Res. 45, W550–W553 (2017).
Ashkenazy, H. et al. Nucleic Acids Res. 44, W344–W350 (2016).
Acknowledgements
We thank V. Olieric for help with S-SAD phasing and data collection at PXIII at the SLS Synchrotron, and the MASSIF beamline team at the ESRF for native data collection. The authors received the following funding support: grant no. NWO 184.032.201 to M.A.; grant nos. NWO 016.Vici.170.033 and KWF NKI-2015–7609, support by the Cancer Genomics Center (CGC.nl) and an Ammodo KNAW Award 2015 to T.R.B.; support by the Oncode Institute to T.R.B. and A.P.
Author information
Authors and Affiliations
Contributions
A.A. performed and analyzed biochemical and structural experiments, wrote a draft manuscript and prepared figures. L.L. performed and analyzed cell-based experiments and prepared figures. T.H. participated in protein purification, crystallization and data collection. F.T. performed expression tests and produced initial crystals, under supervision of A.A. and P.C. O.B.B. and M.A. analyzed the MS samples prepared by J.N. A.P. participated in crystallographic data analysis, wrote the final manuscript and supervised the project together with T.R.B.
Corresponding author
Ethics declarations
Competing interests
T.R.B. is co-founder and SAB member of Haplogen GmbH and co-founder and managing director of Scenic Biotech. All other authors declare no competing interests.
Additional information
Peer review information: Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Conservation of VASH1 and SVBP sequence.
Multiple sequence alignment of orthologous proteins for VASH1 and SVBP. Structural elements are highlighted based on the obtained crystal structure for the VASH1–SVBP complex.
Supplementary Figure 2 Analysis of the VASH1-SVBP interface.
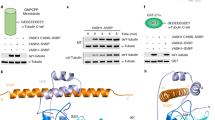
a. Different views of the interacting surface between VASH1 and SVBP; VASH1 is shown as a surface, while SVBP is shown as ribbons; colors are according to conservation of the respective residues based on multiple sequence alignment of orthologous VASH1 proteins, as in Figure 2b. Axes are shown to compare the different views. b. Details for the quantification of detyrosination activity of the SVBP mutants shown in Figure 1e; each individual experiment value is shown as a separate point for the two replicates, and the bar represents the average.
Supplementary Figure 3 Characterization experiments for the VASH1-SVBP structure and the mode of recognition of the tubulin C-terminal peptide.
a. Mutation of a hydrogen bond in the crystallographic dimer of the VASH1–SVBP dimerization interface does not affect detyrosynation activity; detyrosination levels of HeLa cell lysates transfected with vectors expressing Flag-tagged SVBP, VASH1 and mutants were determined using western blotting; this experiments has been performed twice. b. Determination of VASH1 substrate specificity for the α-tubulin peptide based on the number of peptide spectrum matches (PSM) for the intact and cleaved/detyrosinated version of single-residue mutant peptides. c. a representative model from Cluster 1 of the first round of HADDOCK modeling; this model proved wrong on the basis of the validation experiments but is shown here as it was used to guide the first round of validation experiments.
Supplementary Figure 4 Quantification of detyrosination activity of VASH1 mutants.
a-c. The graphs relate to the quantification of activities shown in Figure 3; the bars present the mean and the two circles show the values of the two independent biological replicate experiments. d, e. The Western blot and the quantification establish that a mutation in His80 that was noticed in two of our mutants shown with an asterisk in Figure 3g, does not affect activity compared to the wild type; in the same experiment we confirm again that the K258E and K276E mutants on this background, have no significant effect on activity.
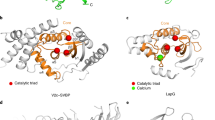
Supplementary Figure 5 Structural analysis of the active site of structural similar proteins to VASH1-SVBP complex.
Ribbon models of structural similar proteins based on DALI server; active site residues for each hit is shown as stick representation in all cases the catalytic triad contains a cysteine and a histidine, whereas the third position is occupied by either a glutamate or an aspartate; PDB codes are: 4DMO for Bacillus cereus arylamine N-acetyltransferase 3 NAT3; 4FGP for LapG, 2PQT for human N-acetyltransferase-1 (the acetanilide molecule that has been covalently attached to the active site C69 is not shown in the figure); 1UKF for Avirulence protein from plant pathogen Pseudomonas syringae; 4RXV for RavJ protein fom Leugionella pneumophila.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Supplementary Tables 1–4 and Supplementary Data Set 1
Rights and permissions
About this article
Cite this article
Adamopoulos, A., Landskron, L., Heidebrecht, T. et al. Crystal structure of the tubulin tyrosine carboxypeptidase complex VASH1–SVBP. Nat Struct Mol Biol 26, 567–570 (2019). https://doi.org/10.1038/s41594-019-0254-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0254-6
This article is cited by
-
The tubulin code and its role in controlling microtubule properties and functions
Nature Reviews Molecular Cell Biology (2020)
-
Cytoskeletal cryptography: structure and mechanism of an eraser
Nature Structural & Molecular Biology (2019)
-
Structural insights into tubulin detyrosination by vasohibins-SVBP complex
Cell Discovery (2019)