Abstract
Wnt signaling plays fundamental roles in organogenesis, tissue regeneration and cancer, but high-resolution structural information of mammalian Wnt proteins is lacking. We solved a 2.8-Å resolution crystal structure of human Wnt3 in complex with mouse Frizzled 8 Cys-rich domain (CRD). Wnt3 grabs the receptor in a manner very similar to that found in Xenopus Wnt8 complexed with the same receptor. Unlike Xenopus Wnt8-bound CRD, however, Wnt3-bound CRD formed a symmetrical dimer in the crystal by exchanging the tip of the unsaturated acyl chain attached to each Wnt3, confirming the ability of Wnt and Frizzled CRD to form a 2:2 complex. The hypervariable ‘linker’ region of Wnt3 formed a β-hairpin protrusion opposite from the Frizzled binding interface, consistent with its proposed role in the coreceptor recognition. Direct binding between this segment and the Wnt coreceptor LRP6 was confirmed, enabling us to build a structural model of the Wnt–Frizzled–LRP6 ternary complex.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Coordinates and structure factors for hWnt3–Fz8 CRD complex have been deposited in the Protein Data Bank (PDB) with the accession number 6AHY. All other data are available from the authors upon request.
References
Logan, C. Y. & Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 (2004).
MacDonald, B. T. & He, X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb. Perspect. Biol. 4, a007880 (2012).
Takada, R. et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell 11, 791–801 (2006).
Willert, K. et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 (2003).
Janda, C. Y., Waghray, D., Levin, A. M., Thomas, C. & Garcia, K. C. Structural basis of Wnt recognition by Frizzled. Science 337, 59–64 (2012).
Chu, M. L. et al. Structural studies of Wnts and identification of an LRP6 binding site. Structure 21, 1235–1242 (2013).
Clevers, H. Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 (2006).
Nile, A. H., Mukund, S., Stanger, K., Wang, W. & Hannoush, R. N. Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding. Proc. Natl Acad. Sci. USA 114, 4147–4152 (2017).
Dann, C. E. et al. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412, 86–90 (2001).
Voss, N. R. & Gerstein, M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 38, W555–W562 (2010).
Fujii, Y. et al. Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state. J. Cell Sci. 129, 1512–1522 (2016).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Mihara, E. et al. Active and water-soluble form of lipidated wnt protein is maintained by a serum glycoprotein afamin/α-albumin. eLife 5, e11621 (2016).
Eubelen, M. et al. A molecular mechanism for Wnt ligand-specific signaling. Science 361, eaat1178 (2018).
Bourhis, E. et al. Reconstitution of a Frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J. Biol. Chem. 285, 9172–9179 (2010).
Zhang, X. et al. Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell 149, 1565–1577 (2012).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013).
Chen, S. et al. Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev. Cell 21, 848–861 (2011).
Zong, Y. et al. Structural basis of agrin-LRP4-MuSK signaling. Genes Dev. 26, 247–258 (2012).
Bourhis, E. et al. Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure 19, 1433–1442 (2011).
Takagi, J., Yang, Y., Liu, J.-H., Wang, J.-H. & Springer, T. A. Complex between nidogen and laminin fragments reveals a paradigmatic β-propeller interface. Nature 424, 969–974 (2003).
Matoba, K. et al. Conformational freedom of the LRP6 ectodomain is regulated by N-glycosylation and the binding of the Wnt antagonist Dkk1. Cell Rep. 18, 32–40 (2017).
Yang, S. et al. Crystal structure of the Frizzled 4 receptor in a ligand-free state. Nature 560, 666–670 (2018).
Chang, T. H. et al. Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. eLife 4, e06554 (2015).
Janda, C. Y. et al. Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545, 234–237 (2017).
Gammons, M. & Bienz, M. Multiprotein complexes governing Wnt signal transduction. Curr. Opin. Cell Biol. 51, 42–49 (2018).
Novarra, S. et al. A hingeless Fc fusion system for site-specific cleavage by IdeS. MAbs 8, 1118–1125 (2016).
von Pawel-Rammingen, U., Johansson, B. P. & Bjorck, L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J. 21, 1607–1615 (2002).
Walter, T. S. et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622 (2006).
Wilkins, M. R. et al. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276, 307–326 (1997).
Mccoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007).
Winn, M. D., Isupov, M. N. & Murshudov, G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57, 122–133 (2001).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Tabata, S. et al. A rapid screening method for cell lines producing singly-tagged recombinant proteins using the “TARGET tag” system. J. Proteomics 73, 1777–1785 (2010).
Acknowledgements
We would like to thank K. Tamura-Kawakami for the construction of expression vectors, and the staff in BL44XU, SPring-8 for their help with X-ray data collection. This work was supported in part by MEXT KAKENHI grant number JP17H01420 from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Innovative Drug Discovery and Life Science Research) funded by Japan Agency for Medical Research and Development under grant number JP18am0101075.
Author information
Authors and Affiliations
Contributions
H.H. designed and performed experiments, analyzed the data and wrote the manuscript. K.M. and E.M. performed experiments, analyzed the data and wrote the manuscript. T.A. supervised the structure determination, analyzed the data and wrote the manuscript. J.T. conceived the experimental design, analyzed the data and wrote the manuscript. All authors contributed to the final editing and approval of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
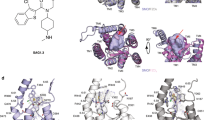
Supplementary Figure 1 Overall structure of Wnt–Fz CRD complex.
a, Structural comparison among the three Wnt proteins. Structures of Xenopus Wnt8/mFz8 CRD complex (PDB ID: 4F0A) and Drosophila WntD N-terminal domain (PDB ID: 4KRR) are shown in ribbon presentation after superposition with the hWnt3–mFz8 CRD structure at NTD, using the same color code as in Fig. 1a. b, Structural superposition of three Wnt proteins. Three Wnt structures are superposed at the core of the NTD comprised by five α-helices (magenta) and shown in Cα tracing models. c, Mobility of Wnt ligands upon the binding to Fz CRD. Three independent hWnt3 structures (chain B, skyblue; chain D, green; chain F, orange) in complex with mFz8 CRD are superposed at the Fz8 CRD (red) and shown as Cα tracings and viewed from two orientations.
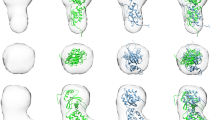
Supplementary Figure 2 Fz–Fz dimer formation.
a, The 2:2 symmetric dimer found in the hWnt3–mFz8 CRD crystal. The upper panel shows three independent hWnt3/mFz8 CRD units (chains AB, CD, and EF) shown with their respective packing mates (chains A’B’, C’D’, and E’F’). The bottom panel shows the crystal structure of apo-form of Fz8 CRD (chains A and B, PDB ID: 1IJY) drawn with their packing mate (chains A’ and B’), PAM-bound Fz5 CRD (chain B, PDB ID: 5URY) drawn with its packing mate (chain B’), as well as a pseudosymmetric dimer of Fz7 CRD bound by a C24 fatty acid (chains A and B, PDB ID: 5URV). The 2-fold symmetry axis is denoted by dotted lines. Lipid molecules are drawn as sphere models. b, Lipophilic cavity at the Fz-Fz interface. The volume of the cavity formed at the Fz-Fz symmetric dimer interface in the 7 dimeric structures in (a) are calculated by using the ChannelFinder program in the 3V server (http://3vee.molmovdb.org/) with the outer and inner probe radius set to 4.0 and 1.4 Å, respectively. The volume map files (.mrc format) produced by the server are opened in UCSF Chimera 1.7.0 and drawn with Fz CRD structures as in (a). The volume of the PAM chain moiety taken from chain B was calculated by the VolumeCalculation program in the same server, resulting in a value of 298 Å3.
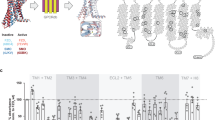
Supplementary Figure 3 Gel filtration profiles of various protein preparations.
a,b, Mouse Wnt3a in complex with mFz8 CRD prepared exactly as hWnt3–mFz8 CRD complex used in the crystallization experiments was loaded to a Superdex 200 gel filtration column at either 200 µM (a, 11 mg/ml) or 9 µM (b, 0.5 mg/ml) and eluted with HBS. The estimated molecular sizes for each peak were calculated from the elution positions of standard marker proteins. c,d, Purity and the monodispersity of the LRP6(3–4) (c) and EGFRec (d) preparations used in the SPR experiments were confirmed by gel filtration on a Superdex 200 column (upper panels) and the SDS-PAGE analysis (lower panels).
Supplementary Figure 4 Fz and Wnt expression constructs used in this study.
Construct design of variously tagged Fz8 (3 versions) and Wnt (12 versions) proteins are schematically shown, with each amino acid sequence of the N-terminal regions starting at the initiation Met indicated at the bottom.
Supplementary Figure 5 Verification of the Wnt3a protein preparations used in the study.
a, SDS-PAGE analysis of the purified afamin-Wnt3a complex sample before and after the biotinylation. Note that the Wnt3a protein used here retains the N-terminal extra 29-residue tag sequence, appearing as larger size than the mature protein (37 kDa). The number of biotin molecules incorporated in one Wnt3a molecule ( = number of Lys modification) was estimated to be 7.35 mol/mol. b, TCF reporter assay to evaluate the signaling activity of the afamin–Wnt3a complex. Unmodified and biotinylated samples were added to the TCF reporter cells at indicated concentrations and the signaling activities were evaluated and expressed as in Fig. 3. c, d, Lack of dissociation of Wnt3a from the surface-captured Fz8 CRD-Fc during the SPR analysis. Shown are the same sensorgrams used to draw Fig. 4d (for c) and e (for d) but are before the Y-axis alignment at the time of injection (time = 190 s). The baseline value at the beginning of each binding cycle (see an expanded view in the inset) remained nearly constant with the total decline of less than 7 RU over one set of experiments, indicating the minimum dissociation of Wnt3a during this period.
Supplementary information
Supplementary information
Supplementary Figs. 1–5, Supplementary Notes 1–3 and Supplementary Dataset 1
Supplementary Video 1
Macroscopic view of the crystal packing. One set of molecules contained in the asymmetric unit are shown with the color code the same as that for Supplementary Fig. 2a, with three more sets that constitute the symmetric packing mate shown in gray.
Supplementary Video 2
Close-up view of the chain A/B pair at the Fz–Fz dimer interface. Fo − Fc electron density map at the tip of Ser 212 is drawn as a magenta mesh at a contour level of σ = 2.5.
Supplementary Video 3
Close-up view of the chain C/D pair at the Fz–Fz dimer interface. Fo – Fc electron density map at the tip of Ser 212 is drawn as a magenta mesh at a contour level of σ = 2.5.
Supplementary Video 4
Close-up view of the chain E/F pair at the Fz–Fz dimer interface. Fo − Fc electron density map at the tip of Ser 212 is drawn as a magenta mesh at a contour level of σ = 2.5.
Rights and permissions
About this article
Cite this article
Hirai, H., Matoba, K., Mihara, E. et al. Crystal structure of a mammalian Wnt–frizzled complex. Nat Struct Mol Biol 26, 372–379 (2019). https://doi.org/10.1038/s41594-019-0216-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0216-z
This article is cited by
-
Chemically-defined and scalable culture system for intestinal stem cells derived from human intestinal organoids
Nature Communications (2024)
-
Frizzled receptors (FZDs) in Wnt signaling: potential therapeutic targets for human cancers
Acta Pharmacologica Sinica (2024)
-
New insights in ubiquitin-dependent Wnt receptor regulation in tumorigenesis
In Vitro Cellular & Developmental Biology - Animal (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Novel WNT10A variant in a Japanese case of nonsyndromic oligodontia
Human Genome Variation (2023)