Abstract
Cas12a is a bacterial RNA-guided nuclease used widely for genome editing and, more recently, as a molecular diagnostic. In bacteria, Cas12a enzymes can be inhibited by bacteriophage-derived proteins, anti-CRISPRs (Acrs), to thwart clustered regularly interspaced short palindromic repeat (CRISPR) adaptive immune systems. How these inhibitors disable Cas12a by preventing programmed DNA cleavage is unknown. We show that three such inhibitors (AcrVA1, AcrVA4 and AcrVA5) block Cas12a activity via functionally distinct mechanisms, including a previously unobserved enzymatic strategy. AcrVA4 and AcrVA5 inhibit recognition of double-stranded DNA (dsDNA), with AcrVA4 driving dimerization of Cas12a. In contrast, AcrVA1 is a multiple-turnover inhibitor that triggers cleavage of the target-recognition sequence of the Cas12a-bound guide RNA to irreversibly inactivate the Cas12a complex. These distinct mechanisms equip bacteriophages with tools to evade CRISPR-Cas12a and support biotechnological applications for which multiple-turnover enzymatic inhibition of Cas12a is desirable.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data for Figs. 1b,d, 3a, 4b and 5a,c are available with the paper online. Source data for all other biochemical experiments that support the findings of this study are available from the corresponding author upon reasonable request. The uncropped images for the main text or Supplementary Figures are available in Supplementary Dataset 1.
References
Wright, A. V., Nuñez, J. K. & Doudna, J. A. Biology and applications of CRISPR systems: harnessing Nature’s toolbox for genome engineering. Cell 164, 29–44 (2016).
Brouns, S. J. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
van Houte, S. et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016).
Pawluk, A., Davidson, A. R. & Maxwell, K. L. Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 16, 12–17 (2018).
Bondy-Denomy, J. Protein inhibitors of CRISPR-Cas9. ACS Chem. Biol. 13, 417–423 (2018).
Koonin, E. V. & Makarova, K. S. Anti-CRISPRs on the march. Science 362, 156–157 (2018).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2012).
Pawluk, A., Bondy-Denomy, J., Cheung, V. H. W., Maxwell, K. L. & Davidson, A. R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 5, e00896-14 (2014).
Landsberger, M. et al. Anti-CRISPR phages cooperate to overcome CRISPR-Cas immunity. Cell 174, 908–916.e12 (2018).
Borges, A. L. et al. Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cell 174, 917–925.e10 (2018).
Pawluk, A. et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016).
Dong, D. et al. Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439 (2017).
Shin, J. et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3, e1701620 (2017).
Harrington, L. B. et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224–1233.e15 (2017).
Watters, K. E., Fellmann, C., Bai, H. B., Ren, S. M. & Doudna, J. A. Systematic discovery of natural CRISPR-Cas12a inhibitors. Science 362, 236–239 (2018).
Marino, N. D. et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242 (2018).
Yamano, T. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016).
Dong, D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016).
Swarts, D. C. & Jinek, M. Cas9 versus Cas12a/Cpf1: structure-function comparisons and implications for genome editing. WIREs RNA 9, e1481 (2018).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016).
Savell, K. E. & Day, J. J. Applications of CRISPR/CAS9 in the mammalian central nervous system. Yale J. Biol. Med. 90, 567–581 (2017).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Strohkendl, I., Saifuddin, F. A., Rybarski, J. R., Finkelstein, I. J. & Russell, R. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell 71, 816–824.e3 (2018).
Swarts, D. C. & Jinek, M. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell 73, 589–600 (2019).
Knott, G. J. & Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Swarts, D. C., van der Oost, J. & Jinek, M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233.e4 (2017).
Stella, S. et al. Conformational activation promotes CRISPR-Cas12a catalysis and resetting of the endonuclease activity. Cell 175, 1856–1871.e21 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Nakamura, M. et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 10, 194 (2019).
Burstein, D. et al. New CRISPR-Cas systems from uncultivated microbes. Nature 542, 237–241 (2017).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Zhao, H., Brown, P. H. & Schuck, P. On the distribution of protein refractive index increments. Biophys. J. 100, 2309–2317 (2011).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Acknowledgements
We thank J. Ye and K. Zhou for technical assistance, and A. Lapinaite, B. Cress and other members of the Doudna laboratory for critical discussions. G.J.K. is a recipient of an American Australian Association Fellowship from the American Australian Association. B.A.S. is supported by the National Science Foundation Graduate Research Fellowship (No. DGE 1752814). This research was developed with funding from Defense Advanced Research Projects Agency (DARPA) award HR0011-17-2-0043 and the Paul G. Allen Frontiers Group, and this material is based on work supported by the National Science Foundation under award number 1817593. J.A.D. is an investigator of the Howard Hughes Medical Institute, and this study was supported in part by that body. J.A.D. is also a Paul Allen Distinguished Investigator.
Author information
Authors and Affiliations
Contributions
G.J.K. conceived the study with input from K.E.W. G.J.K. designed experiments with input from B.W.T., and J.A.D. G.J.K. and B.W.T. carried out biochemical work. M.J.L. and G.J.K. carried out light-scattering experiments. J.L. carried out negative-stain electron microscopy. B.A.S. carried out bioinformatic analysis. G.J.K. drafted the manuscript and all authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The Regents of the University of California have patents pending for CRISPR technologies on which the authors are inventors. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Mammoth Biosciences and Inari. J.A.D. is a Director at Johnson & Johnson and has sponsored research projects by Pfizer and Biogen.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Michaelis–Menten kinetics for the LbCas12a RuvC-nuclease in the absence or presence of AcrVAs.
Representative plots (n=3 independent experiments) of initial velocity versus time (related to main text Fig. 1b) for a) LbCas12a-crRNA, b) LbCas12a-crRNA with 25 nM AcrVA1, c) LbCas12a-crRNA with 25 nM AcrVA4, d) LbCas12a-crRNA with 25 nM AcrVA5. The mean calculated Vmax and Km are reported in a table below for both the 25 nM and 50 nM inhibitor concentrations (mean ± sd, n=3).
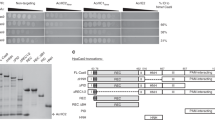
Supplementary Figure 2 Purified Cas12a orthologs and AcrVA used in this study.
a) Unrooted Maximum Likelihood phylogenetic tree of type V effector proteins. Triangle denotes collapsed branches of Cas12b-e. Cas12a orthologs targeted by AcrVA are indicated with circles (closed circles, Cas12a orthologs that are co-occurring with the denoted prophage AcrVA proteins on the same genome; open circles, Cas12a orthologs experimentally inhibited by an AcrVA but without naturally occurring AcrVA orthologs, b) SDS polyacrylamide gel electrophoresis of purified proteins used in this study.
Supplementary Figure 3 AcrVAs differentially affect the cleavage of single-stranded DNA by Cas12a orthologs.
(Top) Representative 5'-radiolabeled kinetic ssDNA cleavage assays (n=3 independent experiments) for a) MbCas12a, b) LbCas12a, and c) AsCas12a in the absence or presence of AcrVAs. Time courses represent 1, 2, 5, 15, and 60 min. The uncleaved and cis-cleaved fractions are indicated with black triangles and the trans-cleavage products are indicated by a black bar. (Bottom) Quantified percentage ssDNA cleaved for a) MbCas12a, b) LbCas12a, and c) AsCas12a in the presence or absence of AcrVAs (mean ± sd, n = 3). Experimental fits are shown as solid lines and the calculated pseudo-first-order rate constants (kobs) (mean ± sd, n=3) are: a) 0.7 ± 0.1 min−1, 1.1 ± 0.1 min−1, 0.04 ± 0.01 min−1, and 0.2 ± 0.08 min−1 for MbCas12a, MbCas12a+AcrVA1, MbCas12a+AcrVA4, and MbCas12a+AcrVA5 respectively, b) 2.6 ± 0.3 min−1, 0.15 ± 0.01 min−1, 0.7 ± 0.06 min−1, and 1.2 ± 0.09 min−1 for LbCas12a, LbCas12a+AcrVA1, LbCas12a+AcrVA4, and LbCas12a+AcrVA5 respectively, c) 4.5 ± 2.8 min−1, 0.08 ± 0.01 min−1, 3.5 ± 1.6 min−1, and 3.1 ± 0.9 min−1 for AsCas12a, AsCas12a+AcrVA1, AsCas12a+AcrVA4, and AsCas12a+AcrVA5 respectively.
Supplementary Figure 4 AcrVAs do not block the binding of ssDNA by dLbCas12a-crRNA.
a) Representative 5'-radiolabeled ssDNA EMSA with increasing concentrations of dLbCas12a-crRNA complexed with or without AcrVA before association with ssDNA (n=3 independent experiments). The bound and unbound fractions are indicated with black triangles, b) Quantified fraction ssDNA bound by dLbCas12a in the presence or absence of AcrVAs determined by EMSA (Extended Data Fig. 4a) (mean ± sd, n = 3). Measured dissociation constants (Kd) are 155 pM ± 20, 229 pM ± 53, 94 pM ± 11, and 371 pM ± 45 in the absence of inhibitor or in the presence of AcrVA1, AcrVA4, or AcrVA5, respectively.
Supplementary Figure 5 Oligomeric state of AcrVA1, AcrVA4 and AcrVA5 in complex with LbCas12a-crRNA.
a) Fig. 2a shown with an adjusted exposure to highlight the presence of a super-shift (red circle). Size exclusion chromatography coupled light scattering traces for b) (top to bottom) LbCas12a-crRNA, apo-AcrVA1, and the LbCas12a-crRNA:AcrVA1 complex, and c) (top to bottom) LbCas12a-crRNA, apo-AcrVA5, and the LbCas12a-crRNA:AcrVA5 complex. Note that the same LbCas12a-crRNA trace is included in the main text and b,c of this figure as a visual reference. The absorbance at 280 nm (blue) and 260 nm (grey) are shown (left axis, milli-absorbance units) with the linear region for the mass estimate corresponding to the relevant peaks (black lines, right axis). The predicted molecular weights (Mwt) for each sample are shown above the graph and the calculated Mwt are indicated above the relevant peak. d) 2D-class averages of LbCas12a-crRNA bound to AcrVA4. The scale bar represents 28 nm.
Supplementary Figure 6 AcrVA4 can displace dsDNA bound to dLbCas12a but not ssDNA in an ATP-independent manner.
a) Representative 5'-radiolabeled dsDNA EMSAs for quantifications presented in Figure 3a (n = 3 independent experiments). A 40 nM effective concentration of dLbCas12a-crRNA was complexed with dsDNA and then titrated against increasing concentrations of AcrVA1, AcrVA4, AcrVA5, or cold dsDNA competitor (left to right), b) 5'-radiolabeled ssDNA EMSA with increasing concentrations of dLbCas12a-crRNA complexed with radiolabeled ssDNA before the addition of (left to right) AcrVA1, AcrVA4, AcrVA5, or cold target strand (TS) ssDNA competitor. c) 5'-radiolabeled dsDNA EMSA with increasing concentrations of dLbCas12a-crRNA complexed with dsDNA in the absence or presence of apyrase or ATP before the addition of AcrVA4. The bound and unbound fractions are indicated with black triangles.
Supplementary Figure 7 AcrVA4 drives the release of TS ssDNA from dLbCas12a-crRNA in the presence of a complementary non-target strand (NTS).
a) (top) Schematic representation for the experimental setup, (bottom) 5'-radiolabeled ssDNA EMSA with increasing concentrations of dLbCas12a-crRNA RNP first complexed with radiolabeled ssDNA before the addition of AcrVA4 and a complementary NTS (0.12 nM, left) or a non-complementary NTS (0.12 nM, right), b) (top) Schematic representation for the experimental setup, (bottom) 5'-radiolabeled dsDNA EMSA with increasing concentrations of dLbCas12a-crRNA first RNP complexed with radiolabeled bubbled dsDNA before the addition of AcrVA4, c) Experimental set up as in panel (a), 5'-radiolabeled ssDNA EMSA with increasing concentrations of dLbCas12a-crRNA RNP first complexed with a radiolabeled ssDNA before the addition of AcrVA4 and a mismatched NTS (0.12 nM), d) Experimental set up as in panel (b), 5'-radiolabeled dsDNA EMSA with increasing concentrations of dLbCas12a-crRNA first complexed with radiolabeled dsDNA before the addition of no competitor (left) or AcrVA4 (right). In all panels the bound and unbound fractions are indicated with black triangles.
Supplementary Figure 8 The effect of AcrVA1 on Cas12a pre-crRNA processing and on crRNA bound by Cas12a.
a) 5'-radiolabeled kinetic pre-crRNA processing assays for (left to right) MbCas12a, LbCas12a, and AsCas12a first complexed with or without AcrVAs. b) 5'-radiolabeled crRNA cleavage for (left to right) MbCas12a, LbCas12a, and AsCas12a first complexed with radiolabeled mature crRNA before addition of AcrVA1. c) 5'-radiolabeled crRNA cleavage for (left to right) MbCas12a-crRNA, LbCas12a-crRNA, and AsCas12a-crRNA first complexed with target ssDNA or dsDNA (1 µM) before the addition of AcrVA1. Black triangles indicate the full-length crRNA, truncated crRNA, or pre-crRNA processed 5'-fragment. Time courses represent 1, 2, 5, 15, and 60 min.
Supplementary Figure 9 Biochemistry of AcrVA1-triggered spacer truncation.
a) 5'-radiolabeled crRNA cleavage with LbCas12a-crRNA complexed in the presence or absence of AcrVA1 for two different crRNA, or b) for five crRNA with different spacer lengths. c) 5'-radiolabeled trans-ssRNA cleavage after 60 minutes for (left to right) trans-ssRNA alone, or trans-ssRNA incubated with AcrVA1, or trans-ssRNA incubated with MbCas12a, LbCas12a, or AsCas12a in the presence of AcrVA1. d) 5'-radiolabeled crRNA cleavage assays with Cas12a-crRNA orthologs complexed without or with AcrVA1 (full-length gel of Fig. 4c). For a single gel, treatments in the absence of AcrVA1 are (left to right) crRNA hydrolysis ladder (OH), crRNA RNase T1 digestion (T1), untreated crRNA (-), and crRNA incubated with Cas12a (Mb/Lb/As). Treatments in the presence of AcrVA1 are (left to right) wild-type Cas12a (Mb/Lb/As), dCas12a (D864A, dMb; D832A, dLb; D908A, dAs), and processing dead Cas12a (K825A, pMb; K785A, pLb; K860A, pAs). A large black triangle indicates full-length crRNA, smaller triangles indicate positions of mapped G-nucleotides for the LbCas12a-crRNA. e) 5'-radiolabeled crRNA binding to LbCas12a shown by EMSA in the absence (left) or presence (right) of AcrVA1, f) 3'-radiolabeled crRNA binding to LbCas12a in the presence of AcrVA1 shown by EMSA. The bound, unbound, or AcrVA1 truncated fractions are indicated with black triangles. g) Quantified fraction of 5'-radiolabeled crRNA cleaved in the presence of LbCas12a and AcrVA1 where the 1X cleavage buffer is modified to have no MgCl2, 5 mM MgCl2, or 25 mM EDTA. Experimental fits are shown as solid lines and the calculated pseudo-first-order rate constants (kobs) (mean ± sd, n=3) are 1.7 ± 0.1 min−1, 1.9 ± 0.2 min−1, and 0.25 ± 0.03 min−1 for conditions containing 0 mM MgCl2, 5 mM MgCl2, or 25 mM EDTA, respectively.
Supplementary Figure 10 Effect of crRNA truncation on ssDNA targeting and the effect of Cas12a domain truncations on crRNA binding/pre-crRNA processing.
a) 5′-radiolabeled kinetic ssDNA cleavage by LbCas12a in the presence of (left to right) crRNA, crRNA and AcrVA1, the 5′-crRNA fragment, 3′-crRNA fragment, or both the 5′- and 3′-crRNA fragments. Final effective crRNA concentration in all reactions was 36 nM. Time courses represent 1, 2, 5, 15, and 60 min. The uncleaved and cis-cleaved fractions are indicated with black triangles and the trans-cleavage products are indicated by a black bar. b) 5′-radiolabeled crRNA EMSA with increasing concentrations of LbCas12a, LbCas12aΔPID (Δ586-677, G4S linker) or LbCas12aΔREC (Δ44-520, G4S linker) complexed with radiolabeled crRNA. The bound and unbound fractions are indicated with black triangles, c) 5′-radiolabeled kinetic pre-crRNA processing assays for LbCas12a, LbCas12aΔPID (Δ586-677, G4S linker) or LbCas12aΔREC (Δ44-520, G4S linker). Time courses represent 1, 2, 5, 15, and 60 min. Black triangles indicate the full-length pre-crRNA or processed crRNA.
Supplementary information
Supplementary Information, Supplementary Table and Supplementary Dataset
Supplementary Figures 1–10, Supplementary Table 1 and Supplementary Dataset 1
Rights and permissions
About this article
Cite this article
Knott, G.J., Thornton, B.W., Lobba, M.J. et al. Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat Struct Mol Biol 26, 315–321 (2019). https://doi.org/10.1038/s41594-019-0208-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0208-z