Abstract
Ubiquitin or ubiquitin-like proteins can be covalently conjugated to multiple proteins that do not necessarily have binding interfaces. Here, we show that an evolutionary transition from covalent conjugation to non-covalent interaction has occurred in the ubiquitin-like autophagy-related 12 (ATG12) conjugation system. ATG12 is covalently conjugated to its sole substrate, ATG5, by a ubiquitylation-like mechanism. However, the apicomplexan parasites Plasmodium and Toxoplasma and some yeast species such as Komagataella phaffii (previously Pichia pastoris) lack the E2-like enzyme ATG10 and the most carboxy (C)-terminal glycine of ATG12, both of which are required for covalent linkage. Instead, ATG12 in these organisms forms a non-covalent complex with ATG5. This non-covalent ATG12–ATG5 complex retains the ability to facilitate ATG8–phosphatidylethanolamine conjugation. These results suggest that ubiquitin-like covalent conjugation can evolve to a simpler non-covalent interaction, most probably when the system has a limited number of targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Uncropped blot images are shown in Supplementary Dataset 1. All other original data are available from the corresponding authors upon reasonable request.
References
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014).
Kwon, Y. T. & Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886 (2017).
van der Veen, A. G. & Ploegh, H. L. Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 (2012).
Vierstra, R. D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160, 2–14 (2012).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Mizushima, N., Sugita, H., Yoshimori, T. & Ohsumi, Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 273, 33889–33892 (1998).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Fujioka, Y. et al. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283, 1921–1928 (2008).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011).
Hanada, T. et al. The ATG12-ATG5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 (2007).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. Mouse Apg10 as an Apg12 conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett. 532, 450–454 (2002).
Cadwell, K. & Debnath, J. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol. 217, 813–822 (2018).
Mizushima, N. & Sahani, M. H. ATG8 localization in apicomplexan parasites: apicoplast and more? Autophagy 10, 1487–1494 (2014).
Besteiro, S. Autophagy in apicomplexan parasites. Curr. Opin. Microbiol. 40, 14–20 (2017).
McFadden, G. I. The apicoplast. Protoplasma 248, 641–650 (2011).
Kong-Hap, M. A. et al. Regulation of ATG8 membrane association by ATG4 in the parasitic protist Toxoplasma gondii. Autophagy 9, 1334–1348 (2013).
Lévêque, M. F. et al. Autophagy-related protein ATG8 has a noncanonical function for apicoplast inheritance in Toxoplasma gondii. mBio 6, e01446–01415 (2015).
Walczak, M., Ganesan, S. M., Niles, J. C. & Yeh, E. ATG8 is essential specifically for an autophagy-independent function in apicoplast biogenesis in blood-stage malaria parasites. mBio 9, e02021–02017 (2018).
Bushell, E. et al. Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272 (2017).
Zhang, M. et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360, eaap7847 (2018).
Shintani, T. et al. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234–5241 (1999).
Suzuki, K. et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 (2001).
Mizushima, N. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–667 (2001).
Kabeya, Y. et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2012 (2004).
Otomo, C., Metlagel, Z., Takaesu, G. & Otomo, T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 20, 59–66 (2013).
Mukaiyama, H. et al. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70 (2004).
Tamura, N., Oku, M. & Sakai, Y. Atg8 regulates vacuolar membrane dynamics in a lipidation-independent manner in Pichia pastoris. J. Cell Sci. 123, 4107–4116 (2010).
Noda, N. N., Fujioka, Y., Hanada, T., Ohsumi, Y. & Inagaki, F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 14, 206–211 (2013).
Yashiroda, H. & Tanaka, K. Hub1 is an essential ubiquitin-like protein without functioning as a typical modifier in fission yeast. Genes Cells 9, 1189–1197 (2004).
Shen, X. X. et al. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 6, 3927–3939 (2016).
Sato, S., Rangachari, K. & Wilson, R. J. Targeting GFP to the malarial mitochondrion. Mol. Biochem. Parasitol. 130, 155–158 (2003).
Nishikawa, Y. et al. Characterisation of Toxoplasma gondii engineered to express mouse interferon-gamma. Int. J. Parasitol. 33, 1525–1535 (2003).
Andenmatten, N. et al. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat. Methods 10, 125–127 (2013).
Mukaiyama, H. et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells 7, 75–90 (2002).
Deitsch, K., Driskill, C. & Wellems, T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 29, 850–853 (2001).
Tamura, N., Oku, M. & Sakai, Y. Atg21 regulates pexophagy via its PI(3)P-binding activity in Pichia pastoris. FEMS Yeast Res. 14, 435–444 (2014).
Gould, S. J., McCollum, D., Spong, A. P., Heyman, J. A. & Subramani, S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast 8, 613–628 (1992).
Sears, I. B., O’Connor, J., Rossanese, O. W. & Glick, B. S. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast 14, 783–790 (1998).
Welter, E., Thumm, M. & Krick, R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy 6, 794–797 (2010).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Acknowledgements
We are grateful to M. Shirakawa for technical assistance. We thank K. Hikosaka for assistance in the culture of P. falciparum, M. Komatsu (Juntendo University) for Atg7+/+ and Atg7-/- MEFs, S. Sato (Universiti Malaysia Sabah) for the pSSPF2 vector, M. Meissner (University of Glasgow) for the P5RT70loxPKillerRedloxPYFP-HX vector, S. Yamaoka (Tokyo Medical and Dental University) for the retroviral pMRXIP vector, T. Kitamura (The University of Tokyo) for the retroviral pMXs-IP vector, and T. Yasui (Osaka University) for the pCG-gag-pol and pCG-VSV-G plasmids. This work was supported by funding from the National Key Research and Development Program of China (No. 2017YFD0500400 to H.J.); The Tokyo Biochemical Research Foundation (to M.H.S.); Grants-in-Aid for Scientific Research on Innovative Areas (No. 25111005, to N.M., and No. 16H0101200, to Y.S.) from the Japan Society for the Promotion of Science; and Exploratory Research for Advanced Technology (ERATO) (No. JPMJER1702, to N.M.) and Core Research for Evolutional Science and Technology (CREST) (No. JPMJCR13M7, to N.N.N.) from the Japan Science and Technology Agency (JST).
Author information
Authors and Affiliations
Contributions
H.J. and N.M. conceived the project. Y.P. and H.J. performed Toxoplasma experiments. J.K.M. and M.H.S performed the Plasmodium experiments under the supervision of K.K. H.S. and H.J. performed the phylogenetic analysis. M.O., Y.S., and H.Y. performed the Komagataella experiments. Y.K. performed mass spectrometry. N.N.N. performed structural analysis. Y.P., H.Y., H.S., J.K.M., and H.J. performed other experiments. Y.P., H.Y., H.S., M.O., J.K.M., Y.S., H.J., and N.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Conservation of ATG10 and ATG12 in Alveolata and Pichiaceae species.
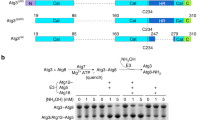
a,b, ATG10 and ATG12 genes and the C-terminal amino acid sequences of ATG12 of representative Alveolata species including apicomplexan parasites (a) and Pichiaceae (b) species are shown. Losses of the ATG10 gene (magenta) and the C-terminal glycine in ATG12 (green) are shown. Black boxes indicate the presence of these genes. n.d., not detected. The residues of ATG12 on a putative binding interface with ATG5 and the C-terminal end are indicated (magenta).
Supplementary Figure 2 Detection of a covalent conjugate and non-covalent complex of Atg12 and Atg5 by mass spectrometry.
a, Schematic representation of mass spectrometry of Atg12 and Atg5 in K. phaffii and S. cerevisiae. Lysates from K. phaffii and S. cerevisiae cells expressing FLAG-KpAtg12 and FLAG-ScAtg12, respectively, were subjected to immunoprecipitation using anti-FLAG antibody. The gels were stained with CBB and three portions at the positions corresponding to the Atg12 monomer, Atg5 monomer, and Atg12–Atg5 conjugate were excised for in-gel trypsin digestion. Trypsin digests from each sample were labeled by stable isotope dimethyl labeling method. Mixtures of labeled peptides were subjected to LC-MS/MS and peptides were identified and quantified. b, List of sequences and quantities of peptides identified. The number indicates the area of the chromatogram of the peptides identified.
Supplementary information
Supplementary Figures and Supplementary Dataset
Supplementary Figures 1–2 and Supplementary Dataset 1
Rights and permissions
About this article
Cite this article
Pang, Y., Yamamoto, H., Sakamoto, H. et al. Evolution from covalent conjugation to non-covalent interaction in the ubiquitin-like ATG12 system. Nat Struct Mol Biol 26, 289–296 (2019). https://doi.org/10.1038/s41594-019-0204-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0204-3
This article is cited by
-
The relationship between autophagy and respiratory viruses
Archives of Microbiology (2024)
-
Autophagy genes in biology and disease
Nature Reviews Genetics (2023)
-
Autophagy and its role in regeneration and remodeling within invertebrate
Cell & Bioscience (2020)
-
Autophagy without conjugation
Nature Structural & Molecular Biology (2019)