Abstract
Replication protein A (RPA) coordinates important DNA metabolic events by stabilizing single-stranded DNA (ssDNA) intermediates, activating the DNA-damage response and handing off ssDNA to the appropriate downstream players. Six DNA-binding domains (DBDs) in RPA promote high-affinity binding to ssDNA yet also allow RPA displacement by lower affinity proteins. We generated fluorescent versions of Saccharomyces cerevisiae RPA and visualized the conformational dynamics of individual DBDs in the context of the full-length protein. We show that both DBD-A and DBD-D rapidly bind to and dissociate from ssDNA while RPA remains bound to ssDNA. The recombination mediator protein Rad52 selectively modulates the dynamics of DBD-D. These findings reveal how RPA-interacting proteins with lower ssDNA binding affinities can access the occluded ssDNA and remodel individual DBDs to replace RPA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data supporting the conclusions of this study are presented in Supplementary Datasets 1–7. Source data for Figs. 1–4, 6, 7 and 9 are available online. Additional data, plasmids for protein expression and code for single-molecule data analysis are available from the corresponding authors upon request.
References
Chen, R. & Wold, M. S. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays 36, 1156–1161 (2014).
Wold, M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997).
Fanning, E., Klimovich, V. & Nager, A. R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 (2006).
Nguyen, B. et al. Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 426, 3246–3261 (2014).
Gibb, B. et al. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS ONE 9, e87922 (2014).
Chen, R., Subramanyam, S., Elcock, A. H., Spies, M. & Wold, M. S. Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res. 44, 5758–5772 (2016).
Arunkumar, A. I., Stauffer, M. E., Bochkareva, E., Bochkarev, A. & Chazin, W. J. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 278, 41077–41082 (2003).
Bochkareva, E., Korolev, S., Lees-Miller, S. P. & Bochkarev, A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 21, 1855–1863 (2002).
Wyka, I. M., Dhar, K., Binz, S. K. & Wold, M. S. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry 42, 12909–12918 (2003).
Bochkareva, E., Frappier, L., Edwards, A. M. & Bochkarev, A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem. 273, 3932–3936 (1998).
Bastin-Shanower, S. A. & Brill, S. J. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 276, 36446–36453 (2001).
Fan, J. & Pavletich, N. P. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 (2012).
Benson, F. E., Baumann, P. & West, S. C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391, 401–404 (1998).
Shinohara, A. & Ogawa, T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391, 404–407 (1998).
New, J. H., Sugiyama, T., Zaitseva, E. & Kowalczykowski, S. C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 (1998).
Sung, P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 (1997).
Sugiyama, T. & Kowalczykowski, S. C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277, 31663–31672 (2002).
Gibb, B. et al. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol. 21, 893–900 (2014).
Pokhrel, N. et al. Monitoring replication protein a (RPA) dynamics in homologous recombination through site-specific incorporation of non-canonical amino acids. Nucleic Acids Res. 45, 9413–9426 (2017).
Brosey, C. A. et al. Functional dynamics in replication protein A DNA binding and protein recruitment domains. Structure 23, 1028–1038 (2015).
Brosey, C. A. et al. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 41, 2313–2327 (2013).
Kumaran, S., Kozlov, A. G. & Lohman, T. M. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry 45, 11958–11973 (2006).
Kolpashchikov, D. M. et al. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 29, 373–379 (2001).
Brosey, C. A. et al. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J. Am. Chem. Soc. 131, 6346–6347 (2009).
Boehm, E. M., Subramanyam, S., Ghoneim, M., Washington, M. T. & Spies, M. Quantifying the assembly of multicomponent molecular machines by single-molecule total internal reflection fluorescence microscopy. Methods Enzymol. 581, 105–145 (2016).
Ghoneim, M. & Spies, M. Direct correlation of DNA binding and single protein domain motion via dual illumination fluorescence microscopy. Nano Lett. 14, 5920–5931 (2014).
Van de Meent, J.-W., Bronson, J. E., Wiggins, C. H. & Gonzalez, R. L. Jr. Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments. Biophys. J. 106, 1327–1337 (2014).
Subramanyam, S., Kinz-Thompson, C. D., Gonzalez, R. L. Jr & Spies, M. Observation and analysis of rad51 nucleation dynamics at single-monomer resolution. Methods Enzymol. 600, 201–232 (2018).
Lu, H. P. & Xie, X. S. Single-molecule spectral fluctuations at room temperature. Nature 385, 143–146 (1997).
Brender, J. R. et al. Conformational dynamics of the isoalloxazine in substrate-free p-hydroxybenzoate hydroxylase: single-molecule studies. J. Am. Chem. Soc. 127, 18171–18178 (2005).
Pretto, D. I. et al. Structural dynamics and single-stranded DNA binding activity of the three N-terminal domains of the large subunit of replication protein A from small angle X-ray scattering. Biochemistry 49, 2880–2889 (2010).
Plate, I. et al. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283, 29077–29085 (2008).
Seong, C. et al. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 283, 12166–12174 (2008).
Sugiyama, T., New, J. H. & Kowalczykowski, S. C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA 95, 6049–6054 (1998).
Hengel, S. R. et al. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. eLife 5, e14740 (2016).
Sugiyama, T. & Kantake, N. Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates. J. Mol. Biol. 390, 45–55 (2009).
Qiu, Y. et al. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun. 4, 2281 (2013).
Subramanyam, S., Ismail, M., Bhattacharya, I. & Spies, M. Tyrosine phosphorylation stimulates activity of human RAD51 recombinase through altered nucleoprotein filament dynamics. Proc. Natl Acad. Sci. USA 113, E6045–E6054 (2016).
Candelli, A. et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl Acad. Sci. USA 111, 15090–15095 (2014).
Chan, H., Wang, Y. & Feigon, J. Progress in human and tetrahymena telomerase structure determination. Annu. Rev. Biophys. 46, 199–225 (2017).
Grimme, J. M. & Spies, M. FRET-based assays to monitor DNA binding and annealing by Rad52 recombination mediator protein. Methods Mol. Biol. 745, 463–483 (2011).
Bain, F. E., Wu, C. G. & Spies, M. Single-molecule sorting of DNA helicases. Methods 108, 14–23 (2016).
Kinz-Thompson, C. D., Bailey, N. A. & Gonzalez, R. L. Jr. Precisely and accurately inferring single-molecule rate constants. Methods Enzymol. 581, 187–225 (2016).
Acknowledgements
We acknowledge the members in our laboratories for their helpful discussions and suggestions. We thank T. Keppel at the Center for Biomedical Mass Spectrometry Research at the Medical College of Wisconsin for MS analysis. This work was supported by grants from the National Institutes of Health (grant no. 7R15GM110671 to E.A. and no. R01 GM108617 to M.S.). C.C.C. is supported by a NIH T32 Pharmacological Sciences training grant (no. NIH T32 GM067795). J.T., S.M.A.T. and M.S. acknowledge support from the University of Iowa Carver College of Medicine FUTURE in Biomedicine program. E.A. also acknowledges support from an SFF-RRG grant from Marquette University. S.M.A.T acknowledges the CHAS Faculty Research Activity grant support from the University of Northern Iowa. E.A. and E.I.C acknowledge salary support from a Department of Energy office of Basic Energy Sciences grant (no. DE-SC0017866).
Author information
Authors and Affiliations
Contributions
N.P., C.C.C., E.I.C., N.J. and E.A.T. performed experiments. J.T. and S.M.A.T. developed the MatLab scripts for data analysis. E.A., M.S., M.S.W., N.P. and C.C.C. conceived and designed the experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 DNA binding properties of fluorescent RPA.
Stopped flow experiments were performed by mixing (a) RPA-wt, (b) RPA-DBD-AMB543 or (c) RPA-DBD-DMB543 with increasing concentrations of ssDNA [(dT)35], and the change in intrinsic tryptophan fluorescence was recorded. (d-f) Data were fit as described in methods to obtain koff and kon values, and the apparent KD values were calculated. (g) Stoichiometric binding of unlabeled or labeled versions of RPA to 32P-labeled [(dT)30] oligonucleotide (50 nM) is observed in EMSA experiments. (h) Occluded site-size measurements were performed by adding increasing concentrations of poly(dT) ssDNA to RPA (0.2 μM) and monitoring the change in tryptophan fluorescence. All versions of RPA occlude ~20 ± 2 nt/RPA in our reaction conditions (30 mM Hepes, pH 7.8, 100 mM KCl, 5 mM MgCl2, 1 mM β-mercaptoethanol and 6 % v/v glycerol). These experiments show that the DNA binding properties of the labeled RPA are similar to the unlabeled wild type RPA complex.
Supplementary Figure 2 Spectral properties of fluorescent RPA and changes in MB543 fluorescence reflect conformational changes associated with electrostatic interactions.
(a) Structure of DBCO-MB543 used to fluorescently label RPA in this study. (b) Excitation and emission spectra of RPA-DBD-AMB543 in solution. (c) Percent change in fluorescence enhancement was measured for RPA-DBD-AMB543 in the absence or presence of ssDNA or (d) dsDNA. The change in fluorescence is specific for ssDNA interactions with RPA. (e) No change in fluorescence is observed for RPA-DBD-ACy5 upon binding to ssDNA. Similar analysis for RPA-DBD-DMB543 were recently published (Nucleic Acids Res. 45, 9413–9426, 2017). (f) The change in MB543 fluorescence was measured as a function of increasing concentrations of dimethyl formamide (DMF) or (h) ethanol, and (g & i) the respective change in fluorescence was plotted. The solvent dependent changes in emission spectra are suggestive of electrostatic changes around the fluorophore (J. Photochem. 3, 55–69, 1974–1975).
Supplementary Figure 3 FRET experiments capture the polarity of RPA binding on ssDNA.
(a) & (c) Models of the expected FRET states and the polarity of the Cy5-DBD with respect to the Cy3-DNA. Boxes are color coded to match the traces in the data. Stopped flow experiments were performed by rapidly mixing either RPA-DBD-ACy5 or RPA-DBD-DCy5 with ssDNA labeled at the (b) 5′ end or (d) 3′ end with Cy3. Cy3 was excited and change in Cy5 emission was measured. The proximity of the Cy3 and Cy5 fluorophores dictate the observed FRET efficiency and results in the enhancement of Cy5 emission. Since RPA binds to ssDNA with a 5′→3′ polarity, when RPA-DBD-ACy5 resides close to the 5′Cy3 on DNA, a high FRET state is observed. Similarly, when RPA-DBD-DCy5 binds close to the 3′Cy3, a high FRET signal is captured. kobs values for change in FRET were obtained by fitting the data to a single exponential plus linear equation. Black traces are DNA only (no RPA). Pink and Green traces are experiments with RPA-DBD-ACy5 and RPA-DBD-DCy5, respectively. The measured rates match well to the rates observed for RPA labeled with MB543 (Fig. 1e, f) suggesting that the ssDNA dependent changes in RPA-MB543 intensity reflect specific DBD-ssDNA interactions.
Supplementary Figure 4 The RPA fragment comprised of FAB domains displays rapid and monophasic binding to ssDNA and single-molecule analysis of RPA-FAB-AMB543-ssDNA interaction reveal the presence of two distinct fluorescence states and more rapid dissociation than trimeric RPA-DBD-AMB543.
The FAB region of RPA (DBDs F, A and B) was purified and labeled with MB543 at DBD-A (RPA-FAB-AMB543), and (a) produces a robust change in fluorescence upon binding to ssDNA. (b) Stopped flow analysis shows rapid binding of RPA-FAB-A MB543 (100 nM) to 100 nM of ssDNA of increasing lengths (dT)n. A minimum of 15 nt is required to observe binding. (c) RPA-FAB-Af binding dynamics on ssDNA were measured by monitoring the change in fluorescence upon binding to increasing concentrations of [(dT)30] ssDNA. (d) Measurement of DNA binding kinetics reveal kon (1.1±0.1 × 108 M-1s-1). The RPA-FAB-AMB543 binding data were fit to a single exponential plus linear equation to obtain kobs,1. (e) Experimental scheme for visualization of the binding and conformational dynamics of FAB. Binding of the fluorescently-labeled FAB (1.0, 0.75, 0.50 µM) to ssDNA (blue line) tethered to the surface of TIRFM flow cell (grey line) brings the MB543 fluorophore within the evanescent field and its excitation. NA – neutravidin, b – biotin. (f) Representative fluorescence trajectories depicting binding (appearance and disappearance of the signal) and conformational dynamics (change in fluorescence without FAB dissociation) of the individual RPA-FAB-AMB543 molecules labeled within the DBD-A. Purple lines represent normalized fluorescence. Black lines represent the results of ebFRET fitting of the experimental data to the three-state model (where state 1 is the off state, while states 2 and 3 are the two bound states with different fluorescent intensities). The levels for the respective states are indicated by dashed lines. The top trajectory is representative of the most commonly observed type with short binding events and rare transitions between fluorescence states. At each RPA-FAB-AMB543 concentration, we also observed several trajectories displaying long binding events as the bottom trajectory here. (g) The dissociation rate constant, koff was determined from the decay rate of the on state dwell time (when trajectories were fitted with the two state model). The average at the three concentrations is shown with standard deviation. (h) The association rate, Von was determined as the number of binding events observed in each experiment divided by the product of the observation time and the number of trajectories observed at 1 nM RPA-FAB-AMB543. The association rate constant, kon was determined by calculating the slope of the Von dependence on RPA-FAB-AMB543 concentration. The equilibrium dissociation rate constant, Kd is the ratio of the two rate constants. All calculations accounted for the RPA-FAB-AMB543 labeling efficiency.
Supplementary Figure 5 Normalization of fluorescence trajectories.
Representative fluorescence trajectory before (a) and after (b) normalization using the emulateFRET program as described in the Methods section. Briefly, to normalize each trajectory we first computed the maximum non-outlying value in this trajectory, which is the 98th percentile value among the values within the trajectory. Each fluorescence value in the trajectory is divided by the maximum non-outlying value; it is further multiplied by a factor of .85 to ensure that the entire trace fits well within the 0–1 range. This resulting trajectory is smoothed with a five-point moving average. Any values in the trace which are still above 1 (these are not expected to occur with any significant frequency) are normalized down to equal 1 (though if this occurs, a small amount of noise is added to these few points, to avoid the trajectory equaling exactly 1 at any point). The second step in normalization determines the location of the new baseline. A histogram of the values from all trajectories collected during the first 30 seconds of the experiment is constructed. This histogram should have a peak at the value at which the baselines center, with a deviation corresponding to the variation in that baseline. The point which is two standard deviations above the mean of this peak is considered the new baseline for the traces. All values in the trace below this value are cut off, and set equal to this new baseline. To avoid the baseline remaining precisely flat (causing over-fitting in ebFRET), a small amount of noise (magnitude .005, at the most) is added throughout the traces. This new set of trajectories is then saved in a format, which ebFRET can read.
Supplementary Figure 6 In the absence of ssDNA, RPA-DBD-DMB543 does not display transitions between fluorescence states; single-molecule analysis of yeast RPA dynamics at several different laser powers shows no trend in number of states, visitation of states, or off-rates at each state, thus ruling out a photophysical cause of fluorescence intensity changes.
To determine whether the four fluorescence states we observed for the ssDNA-bound RPA-DBD-DMB543 are the result of the protein-DNA interaction we tethered RPA-DBD-DMB543 to the surface and observed its fluorescence. (a) Experimental scheme for visualization of RPA-DBD-DMB543 tethered to the surface of the slide chamber via a 6X polyhistidine tag at the C-terminus of RPA32 bound by Biotin-X-NTA (Sigma Cat#51410). b-biotin, N-neutravidin. scRPA tethered to the surface via this immobilization scheme does not bind ssDNA. We believe this is caused by steric constraints of the protein attachment. (b) Representative fluorescence trajectories depicting the fluorescence of tethered RPA-DBD-DMB543. The following rules were followed to ensure that we are analyzing trajectories originating from the single surface-tethered RPA-DBD-DMB543 molecules: selected trajectories (1) showed fluorescence only in the “green” channel and no fluorescence in the “red” channel, (2) terminated in a single-step photobleaching event prior to the last 30 seconds of the movie, and (3) had a signal-to-noise ratio >4. Selected trajectories were normalized similarly to those of the surface-tethered ssDNA except the region of the trajectory after the photobleaching event was used to establish the baseline. Normalized trajectories were fitted with ebFRET to reveal the presence of only two states, fluorescent and the baseline. Photobleaching events in each trajectory are indicated by black arrows while transitions to the dark state are represented by red arrows. The total fluorescent time (7850 frames) was totaled from the trajectories (24) as was the time in the dark state that was not photobleaching (67 frames). 0.85% of time before photobleaching was occupied by dark state fluorescence. Dark state transitions were seen in 5/24 trajectories. These relatively rare dark state transitions can be attributed to blinking. This blinking likely occurs with similarly low frequency in experiments where RPA is bound to ssDNA. (c) In order to verify that the four fluorescent states observed in our experiments were not a result of photophysical effects of the MB543 fluorophore, experiments were carried out at various laser powers. Published work from many groups including ours provides an avenue to distinguish the photophysical source of the fluorescence changes from the actual conformational changes resulting in the change in the dye’s environment 8-10. This is especially important for the low intensity states that can be confused with the dye blinking, such as our state 1. The life times of the photophysical states of the dye should depend on the excitation laser power, while the true conformational states should be independent of it. Blinking frequency is also expected to change with the changing in the power of excitation laser. Experiments were carried out as described in Figure 3b with RPA-DBD-DMB543 in triplicate at three different laser powers (27, 36, and 45mW). Data for each independent experiment is plotted separately. Comparison of the fractional visitation to each state available to RPA-DBD-DMB543 with laser power set to 27 mW (blue), 36 mW (grey), and 45 mW (black). (d) Comparison of the stability of each state available to RPA-DBD-DMB543 at 27 mW (blue), 36 mW (grey), and 45 mW (black). The data on Y axis are the lifetimes calculated from the respective dwell time distributions. Data for each independent experiment is plotted separately. The life times and visitation frequencies for all states were within an experimental error from one laser power to another suggesting that these states are not photophysical.
Supplementary Figure 7 Scavenger ssDNA in the buffer-wash step confirms that RPA-DBD-DMB543 remains bound to ssDNA in observed dark-state events.
In order to verify that the dark fluorescent state that is observed when excess RPA is washed away reflects a dimmer state of RPA-DBD-DMB543 still associated with DNA, we challenged the reaction with excess ssDNA. The free DNA in the reaction chamber is used as a scavenger, which would bind any RPA that dissociated, preventing reassociation, but should not strip the bound RPA from surface-tethered ssDNA5. (a) Experimental scheme for visualization of the effect of excess ssDNA on the RPA-DBD-DMB543 dynamics and association with DNA. Binding of the RPA-DBD-DMB543 (100pM) to the surface-tethered ssDNA (blue line) brings the MB543 fluorophore within the evanescent field and its excitation. 1nM ssDNA (42nt) was added at 90s when excess RPA was washed away. NA – neutravidin, b – biotin. (b) Representative fluorescence trajectories depicting the conformational dynamics of the individual RPA-DBD-DMB543 molecules. After replacement of RPA in the reaction chamber with 1nM ssDNA, the same four conformational states are observed in the RPA-DBD-DMB543 trajectories. As seen in experiments where excess RPA was washed away with reaction buffer, reappearance of higher fluorescence states are observed after visitation of the dark state (state 1) in the presence of excess ssDNA. RPA that dissociated would have been bound by the excess free DNA, thus indicating that the reappearance of higher fluorescence states (2,3,4) is due to RPA remaining bound while in the dark state (1) (c) Comparison of the stability of each state available to RPA-DBD-DMB543 when excess RPA is washed away with reaction buffer (grey) or reaction buffer containing 1nM ssDNA (blue). The data on Y axis are the lifetimes calculated from the respective dwell time distributions. Data for each independent experiment is plotted separately. (d) Comparison of the fractional visitation to each state available RPA-DBD-DMB543 when excess RPA is washed away with reaction buffer (grey) or reaction buffer containing 1nM ssDNA (blue).
Supplementary Figure 8 RPA-FAB is exchanged more readily on ssDNA compared to the full-length RPA.
(a-c) Preformed RPA-FAB-AMB543:(dT)25 complexes were challenged with increasing concentrations of unlabeled RPA-wt and the rate of exchange was measured by monitoring the decrease in fluorescence intensity. RPA-FAB-AMB543 was cleared at an apparent rate of (1.3±0.3x108 M-1s-1). In comparison, (d-h) the rates of exchange of RPA-DBD-AMB543 or RPA-DBD-DMB543 by RPA-wt was two orders of magnitude slower.
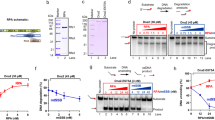
Supplementary Figure 9 The importance of the physical interactions between RPA and Rad52 as well as Rad52 and ssDNA for the modulation of RPA domain dynamics.
The activities of the RAD52 on the RPA-coated ssDNA depend on the species-specific interaction between the two proteins 11. To determine whether the physical interaction between Rad52 and RPA plays a role in the Rad52-mediated change in the RPA conformational dynamics, we carried out the single-molecule analysis of the yeast RPA dynamics in the presence of human RAD52. (a) Experimental scheme for visualization of the effect of hRAD52 on RPA-DBD-D dynamics. Binding of the fluorescently-labeled RPA (100pM) to the ssDNA (blue line) brings the MB543 fluorophore within the evanescent field and its excitation. NA – neutravidin, b – biotin. (b) Human RAD52 protein shares the conserved N-terminal domain with the yeast protein. This domain is responsible for the formation of the oligomeric ring and for the interaction with DNA. Their C-terminal, protein-protein interaction domains are highly different resulting in the absence of cross-species interactions with RPA. (c) Representative fluorescence trajectories depicting the conformational dynamics of the individual RPA molecules labeled within RPA-DBD-D. After replacement of RPA in the reaction chamber with 700pM hRAD52, the same four conformational states are observed in RPA-DBD-DMB543 trajectories suggesting that human RAD52 does not affect the yeast RPA conformational dynamics. Therefore, we conclude that the Rad52-RPA physical interaction is required for the observed effect. (d). Electrophoretic mobility shift assay (EMSA) of Cy5-labeled ssDNA (30nt) contrasting increasing concentrations of scRad52 vs hRAD52 added to scRPA coated ssDNA. Table includes reaction conditions for each lane. Reactions in the bottom portion were crosslinked using 0.1% glutaraldehyde. The species of each band are identified on the right of the gel. The results of this EMSA experiment suggest that the interaction between hRAD52 and scRPA-coated ssDNA are similar to that between scRad52 and the scRPA-coated ssDNA. Thus, the absence of the DBD-D modulation in the single-molecule control experiment is solemnly due to the absence of the protein-protein interaction between the ssDNA-bound scRPA and hRAD52. Previously, we identified epigallocatechin (EGC) as a specific inhibitor of the human RAD52 interaction with ssDNA 12. In contract, EGC displays no activity towards human RPA 12. Here, we confirmed that EGC also inhibits S. cerevisiae Rad52-ssDNA interaction and has no effect on the interaction between S. cerevisiae RPA and ssDNA. We therefore, used EGC to determine whether the interaction between Rad52 and ssDNA is important for the modulation of the RPA conformational dynamics. (e) FRET-based inhibitor titration of EGC into solution of 100 nM Rad52 and 10 nM Cy3-dT30-Cy5 ssDNA. FRET begins high where the FRET labeled DNA is wrapped around Rad52, bringing Cy3 and Cy5 into proximity and decreases as inhibitor prevents Rad52 ssDNA binding. The IC50 value calculated for inhibition of Rad52 ssDNA binding is shown below the curve. (f) FRET-based inhibitor titration of EGC into solution of 10 nM RPA and 10 nM Cy3-dT30-Cy5 ssDNA. The absence of FRET increase with EGC titration indicates that EGC does not inhibit RPA ssDNA binding. (g) Scheme depicting interactions between RPA and Rad52 and DNA. Addition of ECG inhibits Rad52 ssDNA binding. (h) Epigallocatechin (ECG), an inhibitor of Rad52 ssDNA binding. (i) Experimental scheme for visualization of the effect of Rad52 with 10uM EGC on RPA-DBD-D dynamics. Binding of the fluorescently-labeled RPA (100 pM) to the ssDNA (blue line) brings the MB543 fluorophore within the evanescent field and its excitation. NA – neutravidin, b – biotin. (j) Representative fluorescence trajectories depicting the conformational dynamics of the individual RPA molecules labeled within RPA-DBD-D. After replacement of RPA in the reaction chamber with 700 pM Rad52 and 10 uM EGC, the same four conformational states are observed in RPA-DBD-DMB543 trajectories. (k) Comparison of the lifetimes of the individual states and fractional visitation to each state available to RPA-DBD-DMB543 alone (grey) and in the presence of Rad52 (blue) and in the presence of Rad52 and EGC (red). Data from a single experiment was separated into three portions and each portion was analyzed and plotted separately. The presence of EGC returned almost all the lifetimes and all the visitation frequencies to the same values as were obtained in the absence of Rad52. Most importantly, we observed the reemergence of the state 4, suggesting that the Rad52-ssDNA interaction is important for the formation of the RPA-ssDNA-Rad52 complex that modulates the accessibility of the 3′ ssDNA region occluded by RPA. A slight increase in the lifetime of the least fluorescent state 1 is likely the result of the interaction between Rad52 and RPA in the absence of the Rad52-ssDNA interaction.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Notes 1–3, Supplementary Tables 1–4, Supplementary Datasets 1–7
Rights and permissions
About this article
Cite this article
Pokhrel, N., Caldwell, C.C., Corless, E.I. et al. Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat Struct Mol Biol 26, 129–136 (2019). https://doi.org/10.1038/s41594-018-0181-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0181-y
This article is cited by
-
Yeast Rad52 is a homodecamer and possesses BRCA2-like bipartite Rad51 binding modes
Nature Communications (2023)
-
Phase separation properties of RPA combine high-affinity ssDNA binding with dynamic condensate functions at telomeres
Nature Structural & Molecular Biology (2023)
-
ssDNA accessibility of Rad51 is regulated by orchestrating multiple RPA dynamics
Nature Communications (2023)
-
An Aurora B-RPA signaling axis secures chromosome segregation fidelity
Nature Communications (2023)
-
Identification of a small-molecule inhibitor that selectively blocks DNA-binding by Trypanosoma brucei replication protein A1
Nature Communications (2023)