Abstract
Many drugs target the serotonin 2A receptor (5-HT2AR), including second-generation antipsychotics that also target the dopamine D2 receptor (D2R). These drugs often produce severe side effects due to non-selective binding to other aminergic receptors. Here, we report the structures of human 5-HT2AR in complex with the second-generation antipsychotics risperidone and zotepine. These antipsychotics effectively stabilize the inactive conformation by forming direct contacts with the residues at the bottom of the ligand-binding pocket, the movements of which are important for receptor activation. 5-HT2AR is structurally similar to 5-HT2CR but possesses a unique side-extended cavity near the orthosteric binding site. A docking study and mutagenic studies suggest that a highly 5-HT2AR-selective antagonist binds the side-extended cavity. The conformation of the ligand-binding pocket in 5-HT2AR significantly differs around extracellular loops 1 and 2 from that in D2R. These findings are beneficial for the rational design of safer antipsychotics and 5-HT2AR-selective drugs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank under accession codes 6A93 (5-HT2ARris) and 6A94 (5-HT2ARzot). Source data for Figs. 3c and 6c,d and Supplementary Figure 6a,b are available with the paper online. Other data are available from the corresponding authors upon reasonable request.
References
Berger, M., Gray, J. A. & Roth, B. L. The expanded biology of serotonin. Annu. Rev. Med. 60, 355–366 (2009).
Roth, B. L., Willins, D. L., Kristiansen, K. & Kroeze, W. K. 5-Hydroxytryptamine2-family receptors (5-hydroxytryptamine2A, 5-hydroxytryptamine2B, 5-hydroxytryptamine2C): where structure meets function. Pharmacol. Ther. 79, 231–257 (1998).
Nichols, D. E. & Nichols, C. D. Serotonin receptors. Chem. Rev. 108, 1614–1641 (2008).
Branchek, T., Adham, N., Macchi, M., Kao, H. T. & Hartig, P. R. [3H]-DOB(4-bromo-2,5-dimethoxyphenylisopropylamine) and [3H] ketanserin label two affinity states of the cloned human 5-hydroxytryptamine2 receptor. Mol. Pharmacol. 38, 604–609 (1990).
Mestre, T. A., Zurowski, M. & Fox, S. H. 5-Hydroxytryptamine 2A receptor antagonists as potential treatment for psychiatric disorders. Expert Opin. Investig. Drugs 22, 411–421 (2013).
Aznar, S. & Hervig, M. E. The 5-HT2A serotonin receptor in executive function: implications for neuropsychiatric and neurodegenerative diseases. Neurosci. Biobehav. Rev. 64, 63–82 (2016).
Meltzer, H. Y. & Massey, B. W. The role of serotonin receptors in the action of atypical antipsychotic drugs. Curr. Opin. Pharmacol. 11, 59–67 (2011).
Kroeze, W. K. et al. H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology 28, 519–526 (2003).
Roth, B. L., Sheffler, D. J. & Kroeze, W. K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug. Discov. 3, 353–359 (2004).
Muench, J. & Hamer, A. M. Adverse effects of antipsychotic medications. Am. Fam. Physician 81, 617–622 (2010).
Weston-Green, K., Huang, X. F. & Deng, C. Second generation antipsychotic-induced type 2 diabetes: a role for the muscarinic M3 receptor. CNS Drugs 27, 1069–1080 (2013).
Uchiyama, S., Ozaki, Y., Satoh, K., Kondo, K. & Nishimaru, K. Effect of sarpogrelate, a 5-HT2A antagonist, on platelet aggregation in patients with ischemic stroke: clinical-pharmacological dose-response study. Cerebrovasc. Dis. 24, 264–270 (2007).
Meltzer, H. Y. & Roth, B. L. Lorcaserin and pimavanserin: emerging selectivity of serotonin receptor subtype-targeted drugs. J. Clin. Invest. 123, 4986–4991 (2013).
Vanover, K. E. & Davis, R. E. Role of 5-HT2A receptor antagonists in the treatment of insomnia. Nat. Sci. Sleep 2, 139–150 (2010).
Peng, Y. et al. 5-HT2C receptor structures reveal the structural basis of GPCR polypharmacology. Cell 172, 719-+ (2018).
Wacker, D. et al. Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 (2013).
Yin, W. et al. Crystal structure of the human 5-HT1B serotonin receptor bound to an inverse agonist. Cell Discov. 4, 12 (2018).
Wang, S. et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018).
Shimamura, T. et al. Structure of the human histamine H1 receptor complex with doxepin. Nature 475, 65–70 (2011).
Ballesteros, J. A. & Weinstein, H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995).
Isberg, V. et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31 (2015).
Roth, B. L., Lopez, E., Patel, S. & Kroeze, W. K. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? The Neuroscientist 6, 252–262 (2000).
Michino, M. et al. What can crystal structures of aminergic receptors tell us about designing subtype-selective ligands? Pharmacol. Rev. 67, 198–213 (2015).
Mathis, M. V. et al. The US Food and Drug Administration’s perspective on the new antipsychotic pimavanserin. J. Clin. Psychiatry 78, e668–e673 (2017).
Hacksell, U., Burstein, E. S., McFarland, K., Mills, R. G. & Williams, H. On the discovery and development of pimavanserin: a novel drug candidate for Parkinson’s psychosis. Neurochem. Res. 39, 2008–2017 (2014).
Meltzer, H. Y., Matsubara, S. & Lee, J. C. Classification of typical and atypical antipsychotic drugs on the basis of dopamine D-1, D-2 and serotonin2 Pki values. J. Pharmacol. Exp. Ther. 251, 238–246 (1989).
Chu, R. et al. Redesign of a four-helix bundle protein by phage display coupled with proteolysis and structural characterization by NMR and X-ray crystallography. J. Mol. Biol. 323, 253–262 (2002).
Roth, C. B., Hanson, M. A. & Stevens, R. C. Stabilization of the human beta2-adrenergic receptor TM4TM3TM5 helix interface by mutagenesis of Glu122(3.41), a critical residue in GPCR structure. J. Mol. Biol. 376, 1305–1319 (2008).
Yasuda, S. et al. Hot-spot residues to be mutated common in G protein-coupled receptors of class A: identification of thermostabilizing mutations followed by determination of three-dimensional structures for two example receptors. J. Phys. Chem. B 121, 6341–6350 (2017).
Yamashita, K., Hirata, K. & Yamamoto, M. KAMO: towards automated data processing for microcrystals. Acta Crystallog. D Struct. Biol. 74, 441–449 (2018).
Foadi, J. et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 69, 1617–1632 (2013).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
Mccoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Struct. Biol. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Friesner, R. A. et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 (2004).
Inoue, A. et al. TGF alpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat. Methods 9, 1021–1029 (2012).
Schrage, R. et al. The experimental power of FR900359 to study Gq-regulated biological processes. Nat. Commun. 6, 10156 (2015).
Takasaki, J. et al. A novel G alpha(q/11)-selective inhibitor. J. Biol. Chem. 279, 47438–47445 (2004).
Egan, C. T., Herrick-Davis, K. & Teitler, M. Creation of a constitutively activated state of the 5-hydroxytryptamine2A receptor by site-directed mutagenesis: inverse agonist activity of antipsychotic drugs. J. Pharmacol. Exp. Ther. 286, 85–90 (1998).
Cheng, H. C. The power issue: determination of K-B or K-i from IC50—a closer look at the Cheng-Prusoff equation, the Schild plot and related power equations. J. Pharmacol. Toxicol. Methods 46, 61–71 (2001).
Acknowledgements
We thank K. Yamashita and the beamline staff for helping with the data collection at SPring-8. Data were collected at SPring-8 (Proposal nos. 2013A1379, 2013B1184, 2014A1301, 2014B1273, 2015A1044, 2015A1080, 2015B2080 and 2017A2524). This research was supported by the Information Core of the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED), the Research Acceleration Program of the JST (S.I.), JSPS KAKENHI (grant nos. 24370044, 24121715, 26102725, 15H04338, 17K19349, and 18H02388 (T.S.), 26840021 (K.T.K.) and 17K08264 (A.I.)), the PRIME JP17gm5910013 (A.I.) and the LEAP JP17gm0010004 (A.I. and J.A.) from AMED, and the Mitsubishi Foundation (T.S).
Author information
Authors and Affiliations
Contributions
K.T.K. screened, expressed, and purified the constructs, performed crystallization trials for the constructs, collected the data with the help of D.I. and K.H., and performed the docking study. H.A. performed binding assays. D.I. collected and processed the data and solved the structures. C.M. screened, expressed, and purified the constructs, performed crystallization trials, and determined and optimized the crystallization conditions for the constructs. T.A. screened, expressed, and purified the constructs and performed crystallization trials for the constructs. A.I. and F.M.N.K. designed, performed, and analyzed functional assays under the supervision of J.A. N.N. and Y.N. prepared the mutants. T.S. designed and supervised the project. S.I. advised T.S. T.S. and S.I. wrote the manuscript. All authors discussed the results and provided comments regarding the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
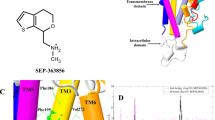
Supplementary Figure 1 Structures of 5-HT2AR antagonists used in this study and electron density maps for the antagonists in the 5-HT2AR crystal structures.
a, Structures of risperidone, zotepine and pimavanserin. |Fo|-|Fc| omit maps (magenta mesh, contoured at 3.0 σ) and polder maps (blue mesh, contoured at 3.0 σ) for risperidone (b) and zotepine (c). The asymmetric unit contains two 5-HT2AR molecules (molecules A and B).
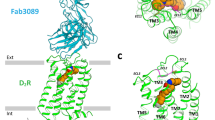
Supplementary Figure 2 Conservation of the bottom hydrophobic cleft in 5-HT2Rs, 5-HT1BR, D2R and H1R.
Vertical cross sections of the receptors. The bottom hydrophobic clefts in the structures are shown in red dotted circles.
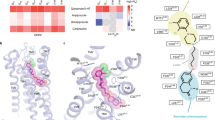
Supplementary Figure 3 The side-extended cavity in 5-HT2Rs.
a, Vertical cross sections of 5-HT2Rs. b, The surface of 5-HT2Rs viewed from the right side of (a). The side chains of Phe2345.38x39 of 5-HT2AR, Phe2175.38x39 of 5-HT2BR and Phe2145.38x39 of 5-HT2CR are shown. 5-HT2AR, 5-HT2BR and 5-HT2CR are shown in pink, orange and green, respectively. Risperidone, ergotamine and ritanserin are shown in magenta, yelloworange and lightgreen, respectively.
Supplementary Figure 4 Comparison of the structures of 5-HT2AR and other aminergic receptors.
a, Superposition of 5-HT2ARzot and the 5-HT1BR-methiothepin structure. Top (b) and bottom (c) views of (a). d, The ligand-binding pocket of (a). 5-HT2AR and 5-HT1BR are shown in cyan and pale green, respectively. Zotepine and methiothepin are shown in green and salmon pink, respectively. e, Superposition of the ligand-binding pockets of 5-HT2ARzot and the H1R-doxepin structure. 5-HT2AR and H1R are shown in cyan and yellow, respectively. Zotepine and doxepin are shown in green and gray-white, respectively.
Supplementary Figure 5 Identification of a Zn ion.
a, The XAFS spectrum of 5-HT2ARris crystals. The yellow dotted line shows the absorption edge of Zn. b, The |Fo|-|Fc| map (blue mesh) and the anomalous difference map (magenta mesh) contoured at 4.0 σ of the data collected at wavelengths of 1.0000 Å (left) and 1.3000 Å (right).
Supplementary Figure 6 TGFα shedding response and the ligand binding assay.
a, Gq/11-dependent TGFα shedding response. The parental HEK293 cells or the Gq/11-deficient HEK293 cells transfected with an empty vector (Mock), WT 5-HT2AR-encoding plasmid or C322K6.34x34 mutant-encoding plasmid were subjected to the TGFα shedding assay in the presence (+) or absence (−) of the Gq/11 inhibitor (1 μM YM-254890). Data represent mean ± SEM from 4–8 independent experiments performed in duplicate or triplicate (see Source data for details). b, The displacement curves of 5-HT2AR-mIIG and the key mutants. The detailed values are shown in Supplementary Table 4. Data represent mean ± SEM from 3 independent experiments performed in triplicate (see Source data for details).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Notes 1–2, Supplementary Tables 1–6
Rights and permissions
About this article
Cite this article
Kimura, K.T., Asada, H., Inoue, A. et al. Structures of the 5-HT2A receptor in complex with the antipsychotics risperidone and zotepine. Nat Struct Mol Biol 26, 121–128 (2019). https://doi.org/10.1038/s41594-018-0180-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0180-z
This article is cited by
-
Pharmacological fingerprint of antipsychotic drugs at the serotonin 5-HT2A receptor
Molecular Psychiatry (2024)
-
Pharmacophore modeling, molecular docking, and molecular dynamics studies to identify new 5-HT2AR antagonists with the potential for design of new atypical antipsychotics
Molecular Diversity (2023)
-
Involvement of the serotonergic system in the antidepressant-like effect of 1-(phenylselanyl)-2-(p-tolyl)indolizine in mice
Psychopharmacology (2023)
-
On the construction of LIECE models for the serotonin receptor 5-HT\(_{\text {2A}}\)R
Journal of Computer-Aided Molecular Design (2023)
-
Structural basis for recognition of antihistamine drug by human histamine receptor
Nature Communications (2022)