Abstract
The majority of eukaryotic proteins are N-terminally α-acetylated by N-terminal acetyltransferases (NATs). Acetylation usually occurs co-translationally and defects have severe consequences. Nevertheless, it is unclear how these enzymes act in concert with the translating ribosome. Here, we report the structure of a native ribosome–NatA complex from Saccharomyces cerevisiae. NatA (comprising Naa10, Naa15 and Naa50) displays a unique mode of ribosome interaction by contacting eukaryotic-specific ribosomal RNA expansion segments in three out of four binding patches. Thereby, NatA is dynamically positioned directly underneath the ribosomal exit tunnel to facilitate modification of the emerging nascent peptide chain. Methionine amino peptidases, but not chaperones or signal recognition particle, would be able to bind concomitantly. This work assigns a function to the hitherto enigmatic ribosomal RNA expansion segments and provides mechanistic insights into co-translational protein maturation by N-terminal acetylation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM map for the ribosome–NatA complex and the respective atomic coordinates have been deposited in the EMDataBank and PDB with accession codes EMD-0201 and 6HD5 for local refinement of NatA, respectively, and with accession codes EMD-0202 and 6HD7 for regular refinement. The map for RNaseI-treated ribosomes was deposited with accession code EMD-0203. Source data for Fig. 3c are available with the paper online. Primary data are available upon reasonable request from the corresponding authors.
References
Gautschi, M. et al. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23, 7403–7414 (2003).
Polevoda, B., Brown, S., Cardillo, T. S., Rigby, S. & Sherman, F. Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J. Cell. Biochem. 103, 492–508 (2008).
Magin, R. S., Deng, S., Zhang, H., Cooperman, B. & Marmorstein, R. Probing the interaction between NatA and the ribosome for co-translational protein acetylation. PLoS ONE 12, e0186278 (2017).
Driessen, H. P., de Jong, W. W., Tesser, G. I. & Bloemendal, H. The mechanism of N-terminal acetylation of proteins. CRC Crit. Rev. Biochem. 18, 281–325 (1985).
Arnesen, T. et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl Acad. Sci. USA 106, 8157–8162 (2009).
Lee, K. E., Heo, J. E., Kim, J. M. & Hwang, C. S. N-terminal acetylation-targeted N-end rule proteolytic system: the Ac/N-end rule pathway. Mol. Cells 39, 169–178 (2016).
Aksnes, H., Drazic, A., Marie, M. & Arnesen, T. First things first: vital protein marks by N-terminal acetyltransferases. Trends. Biochem. Sci. 41, 746–760 (2016).
Drazic, A. et al. NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl Acad. Sci. USA 115, 4399–4404 (2018).
Polevoda, B., Norbeck, J., Takakura, H., Blomberg, A. & Sherman, F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18, 6155–6168 (1999).
Liszczak, G. et al. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 20, 1098–1105 (2013).
Weyer, F. A. et al. Structural basis of HypK regulating N-terminal acetylation by the NatA complex. Nat. Commun. 8, 15726 (2017).
Gottlieb, L. & Marmorstein, R. Structure of human NatA and its regulation by the Huntingtin interacting protein HYPK. Structure, https://doi.org/10.1016/j.str.2018.04.003 (2018).
Varland, S. & Arnesen, T. Investigating the functionality of a ribosome-binding mutant of NAA15 using Saccharomyces cerevisiae. BMC Res. Notes 11, 404 (2018).
Schmidt, C. et al. The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science 354, 1431–1433 (2016).
Schmidt, C. et al. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 44, 1944–1951 (2016).
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001).
Gerbi, S. A. Expansion segment: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA in Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Synthesis (eds. Zimmermann, R. A. & Dahlberg, A. E.) 71–87 (CRC Press, Boca-Raton, FL, USA, 1996).
Hashem, Y. et al. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153, 1108–1119 (2013).
Yamamoto, H. et al. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 34, 3042–3058 (2015).
Bradatsch, B. et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 19, 1234–1241 (2012).
Greber, B. J., Boehringer, D., Montellese, C. & Ban, N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1228–1233 (2012).
Ramesh, M. & Woolford, J. L. Jr. Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA 22, 1153–1162 (2016).
Kater, L. et al. Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171, 1599–1610, e1514 (2017).
Sweeney, R., Chen, L. & Yao, M. C. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol. Cell. Biol. 14, 4203–4215 (1994).
Armache, J. P. et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl Acad. Sci. USA 107, 19748–19753 (2010).
Gomez Ramos, L. M. et al. Yeast rRNA expansion segments: folding and function. J. Mol. Biol. 428, 4048–4059 (2016).
Vetro, J. A. & Chang, Y. H. Yeast methionine aminopeptidase type 1 is ribosome-associated and requires its N-terminal zinc finger domain for normal function in vivo. J. Cell. Biochem. 85, 678–688 (2002).
Sandikci, A. et al. Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat. Struct. Mol. Biol. 20, 843–850 (2013).
Van Damme, P., Hole, K., Gevaert, K. & Arnesen, T. N-terminal acetylome analysis reveals the specificity of Naa50 (Nat5) and suggests a kinetic competition between N-terminal acetyltransferases and methionine aminopeptidases. Proteomics 15, 2436–2446 (2015).
Neuwald, A. F. & Landsman, D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22, 154–155 (1997).
Hou, F., Chu, C. W., Kong, X., Yokomori, K. & Zou, H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J. Cell. Biol. 177, 587–597 (2007).
Evjenth, R. et al. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J. Biol. Chem. 284, 31122–31129 (2009).
Arnesen, T. et al. Cloning and characterization of hNAT5/hSAN: an evolutionarily conserved component of the NatA protein N-alpha-acetyltransferase complex. Gene 371, 291–295 (2006).
Forte, G. M., Pool, M. R. & Stirling, C. J. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 9, e1001073 (2011).
Defenouillere, Q. et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl Acad. Sci. USA 110, 5046–5051 (2013).
Bhushan, S. et al. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell 40, 138–146 (2010).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, https://doi.org/10.7554/eLife.18722 (2016).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Jucker, F. M. & Pardi, A. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry 34, 14416–14427 (1995).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank S. Rieder, J. Musial and H. Sieber for technical assistance. This work was supported by the German Research Council (GRK1721 and FOR1805 to R.B.) and we acknowledge support by the Center for Integrated Protein Science Munich (CiPS-M). A.G.K. is supported by a DFG fellowship through the Graduate School of Quantitative Biosciences Munich.
Author information
Authors and Affiliations
Contributions
B.B., T.B., A.G.K. and R.B. designed the study and wrote the manuscript. A.G.K. purified and biochemically characterized ribosome–NatA complexes and prepared samples for cryo-EM, processed the data, analyzed the structures and, together with T.B., built the molecular model. O.B. collected cryo-EM data. A.G.K. and C.S. processed the cryo-EM data for RNaseI-treated uL4-RNCs. B.B. cloned and established the purification of recombinant NatA. P.T. processed the cryo-EM data for the untreated RNCs.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
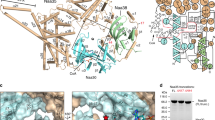
Supplementary Figure 1 Purification of the native ribosome–NatA complex.
a, Schematic representation of the construct for TAP-tagged Naa15. CaMBD, calmodulin-binding domain; TEV, tobacco etch virus protease cleavage site; ProtA, protein A IgG binding domains. b, Amido black–stained PVDF membrane after transfer of purification samples separated on a 4–12% NuPAGE gel. c, Western blot of purification samples probing with an antibody directed against the CaMBD (anti-CAB, Thermo Fisher) and against uL29 (polyclonal chicken antiserum, Davids Biotechnolgie). L, lysate; FT, flow through; R, resuspension; W, wash; E, elution; B, boiled beads. Naa15-CaMBD corresponds to TEV-cleaved product carrying a calmodulin-binding domain, which is recognized by the anti-CAB antibody. 0.1 A260 of E was loaded, for L and FT 1/40,000 and for wash fractions 1/7 of the volume was loaded on the gel.
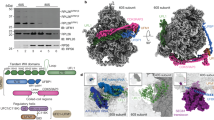
Supplementary Figure 2 3D classification scheme for cryo-EM ribosome–NatA complex.
Particles were picked automatically with Gautomatch, and 2D classification was used to discard non-ribosomal particles. Remaining particles were subjected to 3D refinement and classification. The highest populated classes with ES27a in the exit position were joined and classified according to their tRNA states. Particles containing tRNAs in the A- and P-site were merged and used for further classification with a mask on the ribosomal tunnel exit region. One of the three classes showed well-defined additional density at the exit region for NatA and was refined to an overall resolution of 3.4 Å.
Supplementary Figure 3 Local resolution and FSC curve for the ribosome–NatA complex.
The ribosome–NatA map (see Supplementary Fig. 2) was further refined focusing on the tunnel exit region to improve the resolution of the bound NatA. a,b, Maps before (a) and after (b) focused refinement are shown colored according to their local resolution. Whereas the overall resolution for the ribosome is below 4 Å, NatA displays a higher degree of flexibility indicated by a gradient of local resolution ranging from 7 to >12 Å. Focused refinement on NatA improved the resolution for the ligand to below 10 Å and an average resolution of 8 Å. c,d, FSC curves for the maps shown in a and b, respectively.
Supplementary Figure 4 Docking of NatA subunits into cryo-EM map.
a, Isolated densities for the NatA subunits extracted from the refined map focused on NatA are shown in transparent together with a ribbon representation of the respective molecular models. Naa50 and Naa10 were docked essentially as rigid bodies (derived from PDB 4XNH); Naa15 required some adjustments, especially for the protruding basic helix (see also Supplementary Fig. 7b). b, Zoom views focusing on the ribosome–NatA contacts using the focused refined map as in a. Views were chosen similar to Fig. 2 (see also Supplementary Fig. 3b). c, The upper panel shows the map after global refinement at 3.4 Å with the models for H24, H47 and the N terminus of Naa15. The middle panel shows the same map, but low-pass filtered to 8 Å. Note that density for NatA becomes visible after low-pass filtering. The lower panel shows the map after local refinement low-pass filtered to 8 Å. The density for NatA becomes more defined and distinct interaction sites of NatA with the ribosomal helices become evident.
Supplementary Figure 5 Secondary structure of the 28S rRNA of S. cerevisiae.
Expansion segments ES7a, ES27a and ES39a are labeled in blue. Adopted form http://apollo.chemistry.gatech.edu/RibosomeGallery/
Supplementary Figure 6 Overview of the ribosome-bound NatA structure.
Naa15 is shown in beige, except for the 13 TPR motifs displayed alternating in yellow and blue and numbered according to Liscszak et al. (Nat. Struct. Mol. Biol. 20, 1098–1105, 2013) (PDB 4KVM). TPR11 is not a canonical TPR; instead, it shows extended α-helices and is therefore labeled with 11-like and 11 ext for extended. Naa10 is shown in red and Naa50 is shown in green.
Supplementary Figure 7 Sequence alignment and rearrangement of basic helix α33 of S. cerevisiae and S. pombe.
a, Sequence alignment of the basic helix (α 33) of S. pombe and S. cerevisiae Naa15 with Clustal O (1.2.4). Conserved lysines involved in binding ES39a are boxed. b, Comparison of the free structure of Naa15 of S. pombe (PDB 4KVM) in green with Naa15 of S. cerevisiae in the ribosome-bound conformation in orange. In S. cerevisiae, this basic helix repositions towards ES39a when bound to the ribosome.
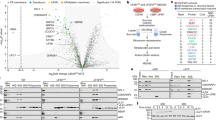
Supplementary Figure 8 Ribosome binding of NatA.
a, Western blot analysis comparing non-treated (RNC) and RNase I–treated (rtRNC) uL4-RNCs. Left, amido black–stained PVDF membrane after transfer of the RNC samples separated on a 12% NuPAGE gel; right, membrane probed with an anti-HA antibody showing the peptidyl-tRNA, indicating that RNase I treatment does not affect the peptidyl-transferase center. Shown is an overlay of the left panel with the antibody signal. b, 4–12% Nu-PAGE gel after an in vitro binding assay showing the supernatant (SN) and pellet (P) fractions of either empty 80S ribosomes or NatA or 80S ribosomes incubated with recombinant NatA. This indicates that NatA is binding to empty ribosomes independently of an emerging nascent chain. c, 15% SDS–PAGE supernatant and pellet fractions of a representative binding assay using either uL4-RNCs (RNCs) or RNase I–treated uL4-RNCs (rtRNCs). Note that significantly less NatA binds to rtRNCs. A representative ribosomal band used for normalization is marked with an asterisk (see also Fig. 3c).
Supplementary Figure 9 Cryo-EM structures of intact and RNase I–treated ribosomes.
Cryo-EM maps of untreated (RNC; orange) and RNase I–treated (rtRNC; gray) ribosomes. The upper panel displays the front view, the middle panel the bottom view and the lower panel a side view. The right column shows a difference map between RNC and rtRNC (red) superimposed on the map of the rtRNCs indicating statistically significant differences between the two maps. Note that mainly density of protruding rRNA is missing in the rtRNCs, such as expansion segments ES7a, ES27a and ES39a or the L1 stalk. Stalk, L7/L12 stalk. All maps were at a resolution of 6.7 Å, low-pass filtered to 8 Å and are shown at the same contour level (3.7σ).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Note
Source data
Rights and permissions
About this article
Cite this article
Knorr, A.G., Schmidt, C., Tesina, P. et al. Ribosome–NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat Struct Mol Biol 26, 35–39 (2019). https://doi.org/10.1038/s41594-018-0165-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0165-y
This article is cited by
-
Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome
Nature Communications (2024)
-
Diverging co-translational protein complex assembly pathways are governed by interface energy distribution
Nature Communications (2024)
-
Structural basis for clearing of ribosome collisions by the RQT complex
Nature Communications (2023)
-
Structure of the actively translating plant 80S ribosome at 2.2 Å resolution
Nature Plants (2023)
-
Structures of the eukaryotic ribosome and its translational states in situ
Nature Communications (2022)