Abstract
The Ku70–Ku80 (Ku) heterodimer binds rapidly and tightly to the ends of DNA double-strand breaks and recruits factors of the non-homologous end-joining (NHEJ) repair pathway through molecular interactions that remain unclear. We have determined crystal structures of the Ku-binding motifs (KBM) of the NHEJ proteins APLF (A-KBM) and XLF (X-KBM) bound to a Ku–DNA complex. The two KBM motifs bind remote sites of the Ku80 α/β domain. The X-KBM occupies an internal pocket formed by an unprecedented large outward rotation of the Ku80 α/β domain. We observe independent recruitment of the APLF-interacting protein XRCC4 and of XLF to laser-irradiated sites via binding of A- and X-KBMs, respectively, to Ku80. Finally, we show that mutation of the X-KBM and A-KBM binding sites in Ku80 compromises both the efficiency and accuracy of end joining and cellular radiosensitivity. A- and X-KBMs may represent two initial anchor points to build the intricate interaction network required for NHEJ.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Crystal structures have been deposited at PDB with the following codes: Ku–DNA–pAPLF (6ERF), Ku–DNA–pXLF (6ERH), and Ku–DNA–pXLFshort (6ERG). Source data for Figs. 3–5 are available with the paper online. Other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Chang, H. H. Y., Pannunzio, N. R., Adachi, N. & Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017).
Walker, J. R., Corpina, R. A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001).
Grundy, G. J., Moulding, H. A., Caldecott, K. W. & Rulten, S. L. One ring to bring them all—The role of Ku in mammalian non-homologous end joining. DNA Repair (Amst.) 17, 30–38 (2014).
Rulten, S. L. & Grundy, G. J. Non-homologous end joining: common interaction sites and exchange of multiple factors in the DNA repair process. Bioessays 39, https://doi.org/10.1002/bies.201600209 (2017).
Costantini, S., Woodbine, L., Andreoli, L., Jeggo, P. A. & Vindigni, A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst.) 6, 712–722 (2007).
Nick McElhinny, S. A., Snowden, C. M., McCarville, J. & Ramsden, D. A. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20, 2996–3003 (2000).
Yano, K. I., Morotomi-Yano, K., Lee, K. J. & Chen, D. J. Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett. 585, 841–846 (2011).
Grundy, G. J. et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 32, 112–125 (2013).
Ropars, V. et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl Acad. Sci. USA 108, 12663–12668 (2011).
Hammel, M. et al. XRCC4 interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double-strand break repair. J. Biol. Chem. 286, 32638–32650 (2011).
Andres, S. N. et al. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 40, 1868–1878 (2012).
Wu, Q. et al. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem. Soc. Trans. 39, 1387–1392 (2011).
Reid, D. A. et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl Acad. Sci. USA 112, E2575–E2584 (2015).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Rulten, S. L., Cortes-Ledesma, F., Guo, L., Iles, N. J. & Caldecott, K. W. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 28, 4620–4628 (2008).
Kanno, S. et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 26, 2094–2103 (2007).
Shirodkar, P., Fenton, A. L., Meng, L. & Koch, C. A. Identification and functional characterization of a Ku-binding motif in Aprataxin Polynucleotide kinase/phosphatase-Like Factor (APLF). J. Biol. Chem. 288, 19604–19613 (2013).
Hammel, M. et al. An intrinsically disordered APLF links Ku, DNA-PKcs, and XRCC4-DNA ligase IV in an extended flexible non-homologous end joining complex. J. Biol. Chem. 291, 26987–27006 (2016).
Arnoult, N. et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 (2017).
Rossi, M. L., Ghosh, A. K. & Bohr, V. A. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst.) 9, 331–344 (2010).
Tadi, S. K. et al. PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep. 17, 541–555 (2016).
Ochi, T. et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347, 185–188 (2015).
Bekker-Jensen, S. et al. Human Xip1 (C2ORF13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 282, 19638–19643 (2007).
Iles, N., Rulten, S., El-Khamisy, S. F. & Caldecott, K. W. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27, 3793–3803 (2007).
Macrae, C. J., McCulloch, R. D., Ylanko, J., Durocher, D. & Koch, C. A. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst.) 7, 292–302 (2008).
Malivert, L. et al. The C-terminal domain of Cernunnos/XLF is dispensable for DNA repair in vivo. Mol. Cell. Biol. 29, 1116–1122 (2009).
Brosey, C. A., Ahmed, Z., Lees-Miller, S. P. & Tainer, J. A. What combined measurements from structures and imaging tell us about DNA damage responses. Methods Enzymol. 592, 417–455 (2017).
Blier, P. R., Griffith, A. J., Craft, J. & Hardin, J. A. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J. Biol. Chem. 268, 7594–7601 (1993).
Dolinsky, T. J. et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007).
Cheng, Q. et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res. 39, 9605–9619 (2011).
Langer, A. et al. Protein analysis by time-resolved measurements with an electro-switchable DNA chip. Nat. Commun. 4, 2099 (2013).
Buck, D. et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124, 287–299 (2006).
Yano, K. I. & Chen, D. J. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle 7, 1321–1325 (2008).
Hammel, M., Yu, Y., Fang, S., Lees-Miller, S. P. & Tainer, J. A. XLF regulates filament architecture of the XRCC4∙ligase IV complex. Structure 18, 1431–1442 (2010).
Malivert, L. et al. Delineation of the XRCC4 interacting region in the globular head domain of cernunnos/XLF. J. Biol. Chem. 285, 26475–26483 (2010).
Bennardo, N., Cheng, A., Huang, N. & Stark, J. M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008).
Bennardo, N., Gunn, A., Cheng, A., Hasty, P. & Stark, J. M. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 5, e1000683 (2009).
Gunn, A., Bennardo, N., Cheng, A. & Stark, J. M. Correct end use during end joining of multiple chromosomal double-strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J. Biol. Chem. 286, 42470–42482 (2011).
Grundy, G. J. et al. The Ku-binding motif is a conserved module for recruitment and stimulation of non-homologous end-joining proteins. Nat. Commun. 7, 11242 (2016).
Brouwer, I. et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–569 (2016).
Andres, S. N., Modesti, M., Tsai, C. J., Chu, G. & Junop, M. S. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell 28, 1093–1101 (2007).
Lu, H., Pannicke, U., Schwarz, K. & Lieber, M. R. Length-dependent binding of human XLF to DNA and stimulation of XRCC4: DNA ligase IV activity. J. Biol. Chem. 282, 11155–11162 (2007).
Wu, P.Y. et al. Structural and functional interaction between the human DNA repair proteins DNA Ligase IV and XRCC4. Mol. Cell Biol. 29, 3163–3172 (2009).
Mari, P. O. et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA 103, 18597–18602 (2006).
Hsu, H. L., Yannone, S. M. & Chen, D. J. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair (Amst.) 1, 225–235 (2002).
Leber, R., Wise, T. W., Mizuta, R. & Meek, K. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 273, 1794–1801 (1998).
Wang, Y. G., Nnakwe, C., Lane, W. S., Modesti, M. & Frank, K. M. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J. Biol. Chem. 279, 37282–37290 (2004).
Cottarel, J. et al. A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol. 200, 173–186 (2013).
Graham, T. G., Walter, J. C. & Loparo, J. J. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell 61, 850–858 (2016).
Menon, V. & Povirk, L. F. XLF/Cernunnos: an important but puzzling participant in the nonhomologous end joining DNA repair pathway. DNA Repair (Amst.) 58, 29–37 (2017).
Jspeert, H. et al. XLF deficiency results in reduced N-nucleotide addition during V(D)J recombination. Blood 128, 650–659 (2016).
Bieniossek, C., Imasaki, T., Takagi, Y. & Berger, I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends. Biochem. Sci. 37, 49–57 (2012).
Kabsch, W. XDS. Acta Crystallogr. D Struct. Biol. 66, 125–132 (2010).
XDS Made Easier; https://github.com/legrandp/xdsme (2018).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Struct. Biol. 67, 235–242 (2011).
Tickle, I. J. et al. STARANISO; http://staraniso.globalphasing.org/ (Global Phasing Ltd., Cambridge, UK; 2018).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D. Struct. Biol. 68, 368–380 (2012).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Struct. Biol. 68, 352–367 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Struct. Biol. 66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Struct. Biol. 66, 12–21 (2010).
Perez, J. & Nishino, Y. Advances in X-ray scattering: from solution SAXS to achievements with coherent beams. Curr. Opin. Struct. Biol. 22, 670–678 (2012).
Petoukhov, M. V. et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 (2012).
Svergun, D. I. Small-angle X-ray and neutron scattering as a tool for structural systems biology. Biol. Chem. 391, 737–743 (2010).
Belin, B. J., Lee, T. & Mullins, R. D. DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-(1/2) that promotes efficient DNA repair. eLife 4, e07735 (2015).
Britton, S., Coates, J. & Jackson, S. P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202, 579–595 (2013).
Huang, F. et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat. Methods 10, 653–658 (2013).
Veatch, S. L. et al. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7, e31457 (2012).
Acknowledgements
J.-B.C. is supported by the ARC program (SLS220120605310), ANR (ANR-12-SVSE8-012), INCA DomRep (PLBIO 2012-280), CEFIPRA grant 5203C, and by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05. Work in E.R.’s laboratory is supported by National Institutes of Health grants CA187612 and GM108119, and by the American Cancer Society RSG DMC-16-241-01-DMC. P.C.’s team is supported by the Ligue Nationale Contre Le Cancer (Equipe labellisée 2013 and 2018) and Electricité de France (EDF, Conseil de Radioprotection). M.M.’s team is supported by the Ligue Nationale Contre Le Cancer. J.-B.C., M.M., and P.C. are supported by ANR (CE12 2017 NHEJLIG4 grant). We thank K.W. Caldecott (University of Sussex, Brighton, UK) for the gift of anti-APLF antibody and J.M. Stark (City of Hope, Duarte, USA) for the gift of U2OS cells engineered with the integrated distal end-joining reporter. We would like to thank the Imaging Core Facility TRI-IPBS, in particular S. Mazeres and R. Poincloux for maintenance of the live-cell microscopy equipment and E. Näser for maintenance of the flow cytometers. Flow cytometry equipment was aquired with financial support from ITMO Cancer Aviesan (Alliance Nationale Pour les Sciences de la Vie et de la Santé, National Alliance for Life Science and Health) within the framework of Cancer Plan. We would also like to thank J.P. de Villartay and F. Theillet for helpful discussions.
Author information
Authors and Affiliations
Contributions
P.C. and J.-B.C. conceived of this study. C.N., V.R., A.G., A.P., A.C., and S.B. expressed and purified recombinant proteins with help from P.D., E.L.C., and I.B. C.N. and V.R. produced crystals and collected crystallographic data with help from P.L. C.N., V.R., P.L., and J.-B.C. carried out the crystallographic analysis and interpreted the results. C.N., A.G., S.B., E.L.C., and J.-B.C. designed, performed, and analyzed microcalorimetry and biophysical experiments. E.B.-M., A.C., and J.-B.C. designed, performed, and analyzed switchSENSE experiments. S.T. and M.M. designed, performed, and analyzed electromobility shift assays. P.F. and P.C. designed and constructed vectors and cell lines for live-cell imaging and radiosensitivity assays. P.F., C.D., N.B., and P.C. designed, performed, and analyzed western blots of Ku variants, live-cell imaging, and DNA repair and radiosensitivity experiments. Y.Y. and E.R. designed, performed, and analyzed the super-resolution microscopy. J.Y. and R.G. performed bioinformatic analyses. All of the authors discussed the data. C.N., P.C., and J.-B.C. wrote the manuscript with input from V.R., P.F., P.D., R.G., M.M., E.B.-M., Y.Y., and E.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
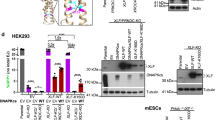
Supplementary Figure 1 Functions and conservation of KBMs.
(a) Scheme of the interactions between Ku70-Ku80 and the NHEJ factors containing an A-KBM (APLF, CYREN), an X-KBM (XLF) and both KBMs (WRN). The interaction of PAXX with Ku70 through its C-terminus is also represented. (b-d) Logo motif of the A-KBM, X-KBM and PAXX motifs obtained from multiple sequences alignment of these proteins as indicated (Crooks, G.E. et al., WebLogo: a sequence logo generator. Genome Res 14, 1188-90 (2004)).
Supplementary Figure 2 Protein purification, structural and ITC data.
(a) SDS gel showing purified Ku and XLF proteins as indicated. FL: full-length; cc: C-terminal truncation. (b) Electron density of peptide pAPLF. (c) Electron density of peptide pXLF. (c) DNA interactions with Ku in presence of X-KBM of XLF. Ku70-Ku80-hDNA-X-KBM (colored) compared to Ku70/Ku80/hDNA (PDB 1JEY, grey). Front view of Ku70-Ku80-hDNA-X-KBM showing the major deviation of hDNA molecules because of the conformational change of Ku80. (d-e) ITC analyses: representative thermograms and isotherms of titration corresponding to selected measurements from Table 2, as indicated.
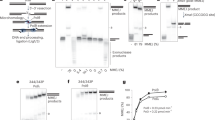
Supplementary Figure 3 EMSA and switchSENSE data.
(a-e) EMSA analyses: (a, b) Gel shift assays with XLF and Ku at 20 nM (a) or 200 nM concentrations (b) in presence of a 50 bp DNA with a FAM in 5′. (c) pXLF interaction with DNA as a control of the competition experiment in Fig. 2e. (d-e) The pAPLF and PAXX Cter do not compete with the Ku-XLF complex. (f-h) switchSENSE analyses: (f) Scheme of the switchSENSE measurement flow: 1) Ku is bound to an 80 bp nanolever with a fluorescent probe at position 48; 2) A washing step removes non-specifically bound Ku molecules; 3) XLF is then injected for real-time associations and dissociations at different concentrations followed by normalized changes in the fluorescence. (g) Binding kinetics of the Ku protein on the 80mer double-stranded DNA prior to the interaction with XLF, shown as changes in the dynamic response upwards (between 0 and 4 µs). The dynamic response reflects the speed of the switching DNA, which decreases upon binding of the Ku analyte. The dissociation is represented only for one minute, to show that no dissociation of Ku from the DNA occurs while the XLF kinetics is measured. (h) Kinetic analyses of (LW) and (L297E)XLF interactions. Solid grey lines represent raw data (from 1 to 8 µM; light grey to dark grey; averages of triplicates). Global fitting was performed, following a single-exponential function (solid orange lines) yielding kinetic rate constants; kON = 4.9 ± 0.5 104 M−1s−1 and kOFF = 4.8 ± 0.5 10−2 s−1 for XLF(L297E) and kON = 1.9 ± 1.1 105 M−1s−1and kOFF = 8.4 ± 0.6 10−2 s−1 for XLF(LW).
Supplementary Figure 4 Evolution of X-KBM binding site in Ku80, comparison with Ku70 and modelling of WRN C terminus binding.
(a) Variations among the sequence motifs observed for the C-terminal tail of XLF and for the seven positions of the Ku80 XBM pocket in various clades of the eukaryotic phylogenetic tree. 10 clades are represented summarizing the properties of 60 Mammalia, 36 Sauria, 31 Neopterygii, 62 Ecdysozoa, 9 Lophotrochozoa, 5 Cnidaria, 150 Fungi, 5 Ciliophora and 4 Dictyostellida sequences of XLF and Ku80. For each clade, web logos of the last 25 C-terminal amino-acids of XLF sequences are represented on top and the web logo of the X-KBM site positions is squared in green. A red star indicates the clades in which the position of Ku80 E133 was conserved as an acidic residue whereas a grey star points out that the acidic character of the residue was not maintained and was generally switched to a hydrophobic residue as observed in Ku70. (b) Superimposition of human Ku70 and Ku80 structures (PDB: 1JEQ) colored in orange and green, respectively, and focused on the region surrounding Ku80 E133 position in the X-KBM site. The red star points out the location of Ku80E133. Residues labelled and shown as sticks are the spatial neighbours of Ku80 E133. The side chain of Ku80 E133 is buried in the hydrophobic core of Ku80 and is not involved in any hydrogen bond or salt-bridge interaction resulting in a predicted pKa above 9.1 in the absence of XLF. (c) Molecular modelling of the interaction between Ku80 and the C-terminus of WRN containing an A-KBM in tandem with a X-KBM. The position of WRN motifs were deduced from the crystal structures presented here with APLF and XLF KBMs. The orientation of the KBMs and the size of the linker between WRN KBMs are compatible with a simultaneous binding of both WRN motifs to Ku80.
Supplementary Figure 5 Western blotting and life cell imaging data.
(a-d) Western blot of U2OS cell extracts. (a) Whole cell extracts of U2OS shKu80 (U2OS-Ku80KD) cells treated with doxycyclin for the indicated time were denatured and separated on 10% SDS-PAGE gel followed by electrotransfer on membrane. The membranes were blotted with the antibodies as indicated. (b) Whole cell extracts of U2OS shAPLF cells expressing WT or mutant CFP-X-KBM as indicated were processed as in (a). (c) Whole cell extracts of U2OS-Ku80KD cells treated with doxycyclin for 7 days and expressing WT or mutant HA-Ku80 as indicated were processed as in (a). (d) Whole cell extracts of U2OS-Ku80KD cells treated with doxycyclin for 7 days and expressing WT or mutant HA-Ku80 as indicated were processed as in (a). Uncropped blot images are shown in Supplementary Data Set 1. (e) Wild-type (WT) or mutant CFP-(A-KBM) and mCherry-Ku70 simultaneous behaviour after 800 nm pulsed-laser nuclear micro-irradiation assessed in U2OS cells by live cell-imaging at 0 s and 50 s post-irradiation. The white rectangle and arrows mark irradiated areas. (f-h) Dynamics of wild-type (WT) and mutant CFP-tagged full-length XLF at laser-damaged sites in BuS cells. Images were obtained at 1.94 s intervals and fluorescence intensities at the damage sites and in undamaged area were quantified. Mean values of the relative fluorescence with SEM were calculated from 20 independent measurements for each of WT and G296W XLF in (f), from 45, 40 and 20 independent measurements for each of WT, S299A and S299E XLF in (g) and from 45, 36, 20 and 20 independent measurements for each of WT, F298G, F298G/S299A, and F298G/S299E XLF in (h), respectively. p values at last time point: (f) WT vs G296W p < 0.0001; (g) WT vs S > A p = 0.2785; WT vs S > E p < 0.0001. (h) WT vs F > G p < 0.0001; WT vs FS > GA p < 0.0001; WT vs FS > GE p < 0.0001. (i) Dynamics of L115D and L115D/L233E CFP-tagged full-length mutant XLF at laser-damaged sites in BuS cells as in (f). Mean values of the relative fluorescence with SEM were calculated from 11 independent measurements for each of L115D ± shAPLF and L115D/L233E XLF conditions. p values at last time point: L115D vs L115D ± shAPLF p = 0.6113; L115D vs L115D/L233E p = 0.0002.
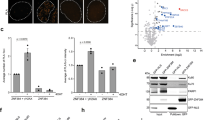
Supplementary Figure 6 Life cell imaging, super resolution and DNA repair assays data.
(a-b) Dynamics of CFP-(A-KBM) (a) and (X-KBM) (b) at laser damaged sites in U2OS cells expressing wild-type (WT), E133M or Q162E mutant Ku80 as in Fig. 3 b). Mean values of the relative fluorescence with SEM were calculated from 20, 23 and 22 independent measurements for A-KBM with WT, E133M or Q162E mutant Ku80 in (a) and from 48, 29 and 29 independent measurements for X-KBM with WT, E133M or Q162E mutant Ku80 in (b), respectively. p values at last time point: (a) WT vs E133M p = 0.831; WT vs Q162E p = 0.59519; (b) WT vs E133M p = 0.0003; WT vs Q162E p = 0.0111. (c-d) Dynamics of wild-type (WT) and mutant CFP-Ku80 at laser damaged sites in U2OS cells. Mean values of the relative fluorescence with SEM were calculated from 25, 24, 20 and 15 independent measurements for WT, I112R, E133M and Q162E mutant Ku80 in (c) and from 25 and 26 independent measurements for WT or I112R/E133M mutant Ku80 in (d), respectively. p values at last time point: (c) WT vs Q162E = 0.9252; WT vs I112R p = 0.2734; WT vs E133M p = 0.1101. (d) WT vs I112R-E1333M p = 0.5362. (e-f) Analysis of XLF foci in U2OS cells by super-resolution. (e) Statistics of XLF foci size: each plot represents the average XLF foci size (indicated as radius translated from the correlation radius) in one nucleus. Box’s height displays the standard deviation with the mean value labelled in the middle. 87, 110, 64, and 79 nuclei were taken in account for WT, E133M, I112R, and E133M-I112R double-mutant, respectively. The p-values were obtained by the t-test; (f) Statistics of the Cross-Pair-Correlation between Ku and XLF: Ku and XLF were stained with antibodies labelled by different fluorophores (Alexa488 conjugated rabbit anti-Ku80, abcam198586, Alexa647 conjugated goat anti-mouse secondary + Mouse anti-XLF, NBP2-03275), and dual-colour super-resolution imaging was performed to examine the cross-correlation between Ku and XLF foci within each nucleus. Each plot represents the cross-correlation amplitude calculated across one nucleus. Box’s height displays the standard deviation with the mean value labelled in the middle. 83, 107, 57, and 72 nuclei were taken in account for WT, E133M, I112R, and E133M-I112R mutants respectively. The p-value were obtained by the t-test.
Supplementary Information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Note
Supplementary Dataset 1
Uncropped western blot images
Rights and permissions
About this article
Cite this article
Nemoz, C., Ropars, V., Frit, P. et al. XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat Struct Mol Biol 25, 971–980 (2018). https://doi.org/10.1038/s41594-018-0133-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0133-6
This article is cited by
-
DNA double-strand break end synapsis by DNA loop extrusion
Nature Communications (2023)
-
Structural basis of long-range to short-range synaptic transition in NHEJ
Nature (2021)
-
The Ku complex: recent advances and emerging roles outside of non-homologous end-joining
Cellular and Molecular Life Sciences (2021)
-
Zinc finger protein ZNF384 is an adaptor of Ku to DNA during classical non-homologous end-joining
Nature Communications (2021)
-
Reply to: “Does PCNA diffusion on DNA follow a rotation-coupled translation mechanism?”
Nature Communications (2020)