Abstract
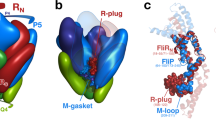
Export of proteins through type III secretion systems is critical for motility and virulence of many major bacterial pathogens. Three putative integral membrane proteins (FliP, FliQ, FliR) are suggested to form the core of an export gate in the inner membrane, but their structure, assembly and location within the final nanomachine remain unclear. Here, we present the cryoelectron microscopy structure of the Salmonella Typhimurium FliP–FliQ–FliR complex at 4.2 Å. None of the subunits adopt canonical integral membrane protein topologies, and common helix-turn-helix structural elements allow them to form a helical assembly with 5:4:1 stoichiometry. Fitting of the structure into reconstructions of intact secretion systems, combined with cross-linking, localize the export gate as a core component of the periplasmic portion of the machinery. This study thereby identifies the export gate as a key element of the secretion channel and implies that it primes the helical architecture of the components assembling downstream.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
17 July 2018
In the version of this article initially published, the PDB code associated with the study was given as 6F2E but should have been 6F2D in Table 1 and the data availability statement. The error has been corrected in the HTML and PDF versions of the article.
23 July 2018
In the version of this paper originally published, the link for Supplementary Video 2 incorrectly led to Supplementary Video 1. This has been corrected in the HTML version of the paper.
References
Abrusci, P., McDowell, M. A., Lea, S. M. & Johnson, S. Building a secreting nanomachine: a structural overview of the T3SS. Curr. Opin. Struct. Biol. 25, 111–117 (2014).
Erhardt, M., Namba, K. & Hughes, K. T. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2, a000299 (2010).
Büttner, D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310 (2012).
Deng, W. et al. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 (2017).
Macnab, R. M. How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100 (2003).
Fabiani, F. D. et al. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol. 15, e2002267 (2017).
Fukumura, T. et al. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 15, e2002281 (2017).
Wagner, S. et al. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. USA 107, 17745–17750 (2010).
Dietsche, T. et al. Structural and functional characterization of the bacterial type III secretion export apparatus. PLoS Pathog. 12, e1006071 (2016).
Zilkenat, S. et al. Determination of the stoichiometry of the complete bacterial type III secretion needle complex using a combined quantitative proteomic approach. Mol. Cell. Proteom. 15, 1598–1609 (2016).
Yonekura, K., Maki-Yonekura, S. & Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 (2003).
Fujii, T. et al. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat. Commun. 8, 14276 (2017).
Matsunami, H., Barker, C. S., Yoon, Y. H., Wolf, M. & Samatey, F. A. Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat. Commun. 7, 13425 (2016).
Cordes, F. S. et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278, 17103–17107 (2003).
Ovchinnikov, S., Kamisetty, H. & Baker, D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife 3, e02030 (2014).
Erhardt, M. et al. Mechanism of type-III protein secretion: regulation of FlhA conformation by a functionally critical charged-residue cluster. Mol. Microbiol. 104, 234–249 (2017).
Ward, E. et al. Type-III secretion pore formed by flagellar protein FliP. Mol. Microbiol. 107, 94–103 (2018).
Worrall, L.J. et al. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540, 597–601 (2016).
Zhao, X. et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 110, 14390–14395 (2013).
Hu, B., Lara-Tejero, M., Kong, Q., Galan, J. E. & Liu, J. In situ molecular architecture of the Salmonella type III secretion machine. Cell 168, 1065–1074.e1010 (2017).
Schraidt, O. & Marlovits, T. C. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331, 1192–1195 (2011).
Minamino, T., Yamaguchi, S. & Macnab, R. M. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182, 3029–3036 (2000).
Hara, N., Namba, K. & Minamino, T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One 6, e22417 (2011).
Radics, J., Königsmaier, L. & Marlovits, T. C. Structure of a pathogenic type 3 secretion system in action. Nat. Struct. Mol. Biol. 21, 82–87 (2014).
Abrusci, P. et al. Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104 (2013).
Chen, S. et al. Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981 (2011).
Minamino, T., Morimoto, Y. V., Hara, N., Aldridge, P. D. & Namba, K. The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog. 12, e1005495 (2016).
Taylor, W. R., Matthews-Palmer, T. R. & Beeby, M. Molecular models for the core components of the flagellar type-III secretion complex. PLoS One 11, e0164047 (2016).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44 (W1), W344–W350 (2016).
Hoiseth, S. K. & Stocker, B. A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981).
Baumgarten, T. et al. Isolation and characterization of the E. coli membrane protein production strain Mutant56(DE3). Sci. Rep. 7, 45089 (2017).
Gault, J. et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods 13, 333–336 (2016).
Marty, M. T., Hoi, K. K., Gault, J. & Robinson, C. V. Probing the lipid annular belt by gas-phase dissociation of membrane proteins in nanodiscs. Angew. Chem. Int. Edn. Engl. 55, 550–554 (2016).
Reboul, C. F., Eager, M., Elmlund, D. & Elmlund, H. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci. 27, 51–61 (2018).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Urnavicius, L. et al. The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446 (2015).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 (2016).
Swint-Kruse, L. & Brown, C. S. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D. Biol. Crystallogr. 71, 136–153 (2015).
Ovchinnikov, S. et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife 4, e09248 (2015).
Monjarás Feria, J. V., Lefebre, M. D., Stierhof, Y. D., Galán, J. E. & Wagner, S. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. MBio 6, e01459–e15 (2015).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Acknowledgements
We thank E. Johnson and A. Costin of the Central Oxford Structural Microscopy and Imaging Centre and all staff of the ARC Centre of Advanced Imaging, Monash University, for assistance with data collection. We thank H. Elmlund (Monash) for assistance with data collection, processing strategies and access to SIMPLE code ahead of release. For collection of preliminary data, we acknowledge Diamond for access and support of the Cryo-EM facilities at the UK National Electron Bio-Imaging Centre (eBIC), proposal EM15354, funded by the Wellcome Trust, MRC and BBSRC. We thank T. Marlovits, Centre for Structural Systems Biology, Hamburg, for providing PrgK (SctJ) antisera. The Central Oxford Structural Microscopy and Imaging Centre is supported by the Wellcome Trust (201536), The EPA Cephalosporin Trust, the Wolfson Foundation and a Royal Society/Wolfson Foundation Laboratory Refurbishment Grant (WL160052). Work performed in the laboratory of S.M.L. was supported by a Wellcome Trust Investigator Award (100298) and an MRC programme grant (M011984). Work performed in the lab of C.V.R. is funded by a Wellcome Trust Investigator Award (104633), an ERC Advanced Grant ENABLE (641317) and an MRC programme grant (N020413). J.G. is a Junior Research Fellow of The Queen’s College, Oxford. Work performed in the laboratory of S.W. was supported by the Alexander von Humboldt Foundation in the framework of the Sofja Kovalevskaja Award endowed by the Federal Ministry of Education and Research (BMBF) and by the Deutsche Forschungsgemeinschaft (DFG) as part of the Collaborative Research Center (SFB) 766 Bacterial Cell Envelope, project B14.
Author information
Authors and Affiliations
Contributions
All authors contributed to editing of manuscript and figures. L.K. performed experiments: strain and plasmid construction, complex purification, nMS, cryo-EM grid optimization, cryo-EM data analysis and model building and analysis. P.A. performed experiments: strain and plasmid construction, complex purification, initial cryo-EM grid optimization and cryo-EM data analysis. S.J. performed experiments and designed and supervised experiments for characterization of protein complexes, cryo-EM data analysis, structure determination and analysis, and wrote the first draft of manuscript and figures with S.M.L. J.G. performed and supervised nMS experiments. J.D. performed experiments: cryo-EM grid optimization and data collection. J.C. performed experiments: optimized computational analysis of cryo-EM data. T.D. performed experiments: strain and plasmid construction, in vivo photo-cross-linking. M.T.M. performed experiments: NC purification, chemical cross-linking. T.G. performed experiments for nMS of DSS cross-linked samples and analyzed data. B.M. analyzed DSS nMS data and wrote the DSS nMS methods section. S.W. analyzed data for the in vivo photo-cross-linking, chemical cross-linking and DSS nMS, and wrote the first draft of photo- and chemical-cross-linking sections. C.V.R. supervised design and implementation of nMS experiments. S.M.L. performed and supervised experiments for cryo-EM grid optimization and data collection, data and structure analysis, and wrote the first draft of paper with S.J.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Characterization of the S. Typhimurium flagellar PQR.
(a) High energy dissociation spectrum of the FliPQR complex from S. Typhimurium extracted, purified and sprayed in DDM detergent. nMS followed by high energy collisional activation (200 V HCD, 150 V in source dissociation) causes dissociation of the PQR assembly and formation of many different subcomplexes containing combinations of P and R subunits. The extracted subunit spectra are displayed in color below the raw data after processing in Unidec. No subcomplexes containing Q could be observed implying it is less tightly associated. This suggests that R is integrated into the PR core complex and Q is more peripherally bound. (b) SEC-MALS analysis on a Superdex200 column of FliPQR purified in LMNG co-expressed with FlhB and FlhA. Molecular weight of the protein component across the peak is represented as a red line and the portion of the peak used for cryo-EM is highlighted in purple. The inset shows SDS-PAGE of the sample confirming the presence of all three components plus FlhB (levels of which vary across the peak) which is seen in two fragments in the gel annotated FlhBN and FlhBC for N and C terminal fragments respectively.
Supplementary Figure 2 Single-particle cryo-EM of FliPQR in LMNG.
(a) Representative micrograph of the FliPQR complex. (b) Selected reference-free 2D class averages. (c) Gold-standard FSC curve of the final map calculated using a soft-edged mask. (d) Local resolution estimates of the final map calculated using ResMap (Swint-Kruse & Brown, 2005). On the left is the overall map with representative slices through the map shown on the right. (e) Examples of good (left), average (middle) and bad (right) density.
Supplementary Figure 3
Flow chart summarizing the processing of the Titan Krios dataset.
Supplementary Figure 4 Purification and crosslinking of the S. Typhimurium T3SS-1 needle complex base.
(a) Negative stain electron micrograph of purified bases of an S. Typhimurium strain with an enzymatically inactive InvC (SctN) ATPase. (b) Coomassie-stained SDS PAGE gel showing the protein content of purified bases with and without crosslinking by DSS. (c) Coomassie-stained SDS PAGE gel showing the briefly run sample (same as in (b)) that was used for in-gel digestion and mass spectrometry. (d) Secretion assay showing functionality of PrgK (SctJ)-pBpa mutants used for in vivo photocrosslinking. Type III-dependent secretion into the culture supernatant of the wild type and indicated pBpa mutants of PrgK (SctJ) was assayed by SDS PAGE and immunodetection of the early substrate InvJ and the intermediate substrate SipB. (e) Uncropped originals of Western blots shown in Fig. 4b. In vivo photo-crosslinking studies reveal cross-links between P5Q4R1 and the inner membrane ring component SctJ in the S. Typhimurium injectisome. SctJ tends to crosslink pBpa-indendent to some extent. * indicates these pBpa-independent crosslinks. SctJE138X crosslinks readily to neighbouring SctJE138X. This results in a crosslink-ladder, of which the homodimeric SctJ-SctJ crosslink is indicated by **. Consequently, also SctJ-SctJ-RFLAG crosslinks are readily detected. Due to the less focussed running behavior of SctJ-PFLAG crosslinks on the SDS-gel, SctJ-SctJ-PFLAG crosslinks are less clearly apparent. *** indicates a band likely resulting from a SctJ-Q crosslink. Q is 9 kDa. Since there is no reliable antibody against Q available and since tagging of Q results in loss of function of the protein, a reciprocal validation of this crosslink is not possible at this point. The images result from a dual-color detection using the LiCor Odyssey system. Therefore, the all blue molecular mass marker is only visible in the 700 nm emission channel that was used for detecting the polyclonal rabbit anti-SctJ (PrgK) antibody.
Supplementary Figure 5 Annotated MS/MS spectra for the QYGEETETVKR (SpaP)–VAKDDSLR (InvG) cross-linked peptide.
Annotated MS/MS spectra for both heavy (a) and light (b) crosslinks. Light (595.8058 m/z) and heavy (598.8228 m/z) DSS crosslinked peptides appear as a quadruply charged doublet with the expected 12.075 Da mass shift (c).
Supplementary Figure 6 Annotated MS/MS spectra for the DKDEIEKPSIF (SpaP)–VAKDDSLR (InvG) cross-linked peptide.
Annotated MS/MS spectra for both heavy (a) and light (b) crosslinks. Light (591.0604 m/z) and heavy (594.0784 m/z) DSS crosslinked peptides appear as a quadruply charged doublet with the expected 12.075 Da mass shift (c).
Supplementary Figure 7 Alignment of typical flagellar and non-flagellar sequences of P, Q and R annotated with secondary structure as seen in our 3D model.
FliPQR and SctRST sequences are those of S. Typhimurium.
Supplementary Figure 8 Reinterpretation of earlier tomograms in light of the structure of PQR. Figure taken from Hu et al, 2017.
(a) Tomogram of wild type S. Typhimurium T3SS. PQR complex is positioned within the image and yellow highlights probable course of inner membrane whilst green denotes density likely contributed by cytoplasmic and membrane domains of A. (b) Tomogram of S. Typhimurium T3SS from a strain lacking expression of A. The PQR complex is shown and yellow indicates the inner membrane which seems distorted to contact the base of the PQR complex in the absence of A.
Supplementary Information
Supplementary Figures
Supplementary Figures 1–8 and Supplementary Tables 1–5
Supplementary Data Set 1
Native mass spectroscopy files used to generate Fig. 1
Supplementary Video 1
Animation of fitting of Gremlin models into the experimental cryo-EM map. The movie starts by showing the fitting of the FliP(1) in the unsharpened experimental map. After morphing to the final protein model, the map is replaced by the final sharpened map (see Methods) and side chains are shown. The zoom is then reset and the fitting of the other chains shown in the unsharpened map
Supplementary Video 2
Morph between closed and open states of the PQR complex. The PQR complex is shown in the context of the coordinates for the rest of the S. Typhimurium injectisome basal body (PDB 5TCR). The closed state is the structure seen by cryo-EM; the open state is modeled on the basis of the initial Gremlin models as described in the Methods
Rights and permissions
About this article
Cite this article
Kuhlen, L., Abrusci, P., Johnson, S. et al. Structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 25, 583–590 (2018). https://doi.org/10.1038/s41594-018-0086-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0086-9
This article is cited by
-
Cytosolic sorting platform complexes shuttle type III secretion system effectors to the injectisome in Yersinia enterocolitica
Nature Microbiology (2024)
-
An overview of the structure and function of the flagellar hook FlgE protein
World Journal of Microbiology and Biotechnology (2023)
-
Evolutionary Conservation, Variability, and Adaptation of Type III Secretion Systems
The Journal of Membrane Biology (2022)
-
Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nature Communications (2021)
-
The FlgN chaperone activates the Na+-driven engine of the Salmonella flagellar protein export apparatus
Communications Biology (2021)