Abstract
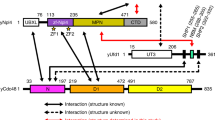
Many polyubiquitinated proteins are extracted from membranes or complexes by the conserved ATPase Cdc48 (in yeast; p97 or VCP in mammals) before proteasomal degradation. Each Cdc48 hexamer contains two stacked ATPase rings (D1 and D2) and six N-terminal (N) domains. Cdc48 binds various cofactors, including the Ufd1–Npl4 heterodimer. Here, we report structures of the Cdc48–Ufd1–Npl4 complex from Chaetomium thermophilum. Npl4 interacts through its UBX-like domain with a Cdc48 N domain, and it uses two Zn2+-finger domains to anchor the enzymatically inactive Mpr1–Pad1 N-terminal (MPN) domain, homologous to domains found in several isopeptidases, to the top of the D1 ATPase ring. The MPN domain of Npl4 is located above Cdc48’s central pore, a position similar to the MPN domain from deubiquitinase Rpn11 in the proteasome. Our results indicate that Npl4 is unique among Cdc48 cofactors and suggest a mechanism for binding and translocation of polyubiquitinated substrates into the ATPase.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stach, L. & Freemont, P. S. The AAA+ATPase p97, a cellular multitool. Biochem. J. 474, 2953–2976 (2017).
Zhang, X. et al. Structure of the AAA ATPase p97. Mol. Cell 6, 1473–1484 (2000).
Tang, W. K. et al. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 29, 2217–2229 (2010).
Banerjee, S. et al. 2.3Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 351, 871–875 (2016).
H„nzelmann, P. & Schindelin, H. The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA+ATPase p97. Front. Mol. Biosci. 4, 21 (2017).
Meyer, H. H., Shorter, J. G., Seemann, J., Pappin, D. & Warren, G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192 (2000).
Ye, Y., Meyer, H. H. & Rapoport, T. A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 (2001).
Skrott, Z. et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199 (2017).
Isaacson, R. L. et al. Detailed structural insights into the p97-Npl4-Ufd1 interface. J. Biol. Chem. 282, 21361–21369 (2007).
McCullough, J., Clague, M. J. & Urbé, S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell. Biol. 166, 487–492 (2004).
Cope, G. A. et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 (2002).
Maytal-Kivity, V., Reis, N., Hofmann, K. & Glickman, M. H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3, 28 (2002).
Alam, S. L. et al. Ubiquitin interactions of NZF zinc fingers. EMBO J. 23, 1411–1421 (2004).
Hänzelmann, P. & Schindelin, H. Characterization of an additional binding surface on the p97 N-terminal domain involved in bipartite cofactor interactions. Structure 24, 140–147 (2016).
Hetzer, M. et al. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3, 1086–1091 (2001).
Heo, J.-M. et al. A stress-responsive system for mitochondrial protein degradation. Mol. Cell 40, 465–480 (2010).
Bodnar, N. O. & Rapoport, T. A. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735.e9 (2017).
Lee, C., Prakash, S. & Matouschek, A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J. Biol. Chem. 277, 34760–34765 (2002).
Burton, R. E., Siddiqui, S. M., Kim, Y. I., Baker, T. A. & Sauer, R. T. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 20, 3092–3100 (2001).
Park, S., Isaacson, R., Kim, H. T., Silver, P. A. & Wagner, G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 13, 995–1005 (2005).
DeLaBarre, B., Christianson, J. C., Kopito, R. R. & Brunger, A. T. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell 22, 451–462 (2006).
Prakash, S., Tian, L., Ratliff, K. S., Lehotzky, R. E. & Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 (2004).
Pye, V. E. et al. Structural insights into the p97-Ufd1-Npl4 complex. Proc. Natl Acad. Sci. U. S. A. 104, 467–472 (2007).
Bebeacua, C. et al. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proc. Natl Acad. Sci. U. S. A. 109, 1098–1103 (2012).
Bruderer, R. M., Brasseur, C. & Meyer, H. H. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J. Biol. Chem. 279, 49609–49616 (2004).
Blok, N. B. et al. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc. Natl Acad. Sci. U. S. A. 112, E4017–E4025 (2015).
Ambroggio, X. I., Rees, D. C. & Deshaies, R. J. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2, E2 (2004).
Lander, G. C. et al. Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 (2012).
Worden, E. J., Dong, K. C. & Martin, A. An AAA motor-driven mechanical switch in Rpn11 controls deubiquitination at the 26 S proteasome. Mol. Cell 67, 799–811.e8 (2017).
Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 (2008).
Davies, C. W., Paul, L. N., Kim, M.-I. & Das, C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J. Mol. Biol. 413, 416–429 (2011).
Tsuchiya, H. et al. In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 Axis contributes to K48-linked chain specificity of the proteasome. Mol. Cell 66, 488–502.e7 (2017).
Worden, E. J., Padovani, C. & Martin, A. Structure of the Rpn11–Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 21, 220–227 (2014).
Shi, Y. et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351, aad9421 (2016).
Matyskiela, M. E., Lander, G. C. & Martin, A. Conformational switching of the 26 S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 (2013).
Chen, S. et al. Structural basis for dynamic regulation of the human 26 S proteasome. Proc. Natl Acad. Sci. U. S. A. 113, 12991–12996 (2016).
Frey, S. & Görlich, D. A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A. 1337, 95–105 (2014).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004).
Li, X., Zheng, S., Agard, D. A. & Cheng, Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Rohou, A. & Grigorieff, N. CTFFIND4: FAST and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Yang, Z., Fang, J., Chittuluru, J., Asturias, F. J. & Penczek, P. A. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 (2012).
Hohn, M. et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Pettersen, E. F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Kabsch, W. X. D. S. Acta Crystallogr. D. Biol. Crystallogr 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D. Biol. Crystallogr. 69, 1204–1214 (2013).
Jackson, R. N., McCoy, A. J., Terwilliger, T. C., Read, R. J. & Wiedenheft, B. X-ray structure determination using low-resolution electron microscopy maps for molecular replacement. Nat. Protoc. 10, 1275–1284 (2015).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D. Biol. Crystallogr. 56, 965–972 (2000).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 (2011).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. Biol. Crystallogr. 64, 61–69 (2008).
Touw, W. G., van Beusekom, B., Evers, J. M. G., Vriend, G. & Joosten, R. P. Validation and correction of Zn-CysxHisy complexes. Acta Crystallogr. D. Struct. Biol. 72, 1110–1118 (2016).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Zubarev, R. A. & Makarov, A. Orbitrap mass spectrometry. Anal. Chem. 85, 5288–5296 (2013).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Fan, S.-B. et al. Using pLink to analyze cross-linked peptides. Curr. Protoc. Bioinforma. 49, 8.21 (2015).
Wang, L., Pan, H. & Smith, D. L. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol. Cell. Proteom. 1, 132–138 (2002).
Wales, T. E., Fadgen, K. E., Gerhardt, G. C. & Engen, J. R. High-speed and high-resolution UPLC separation at zero degrees celsius. Anal. Chem. 80, 6815–6820 (2008).
Wales, T. E. & Engen, J. R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass. Spectrom. Rev. 25, 158–170 (2006).
DeHoratius, C. & Silver, P. A. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell. 7, 1835–1855 (1996).
Acknowledgements
We thank X. Wu and L. Li for assistance with crystallography, D. Finley for critical reading of the manuscript, the SBGrid consortium at Harvard Medical School, and the ICCB Longwood for use of equipment. We thank M. Ebrahim and J. Sotiris at the Rockefeller University Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center for assistance with microscope operation. This work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the NIH/NIGMS (P41 GM103403). The Pilatus 6 M detector on the 24-ID-C beamline is funded by an NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. Z.J. is supported as a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation, DRG-2315-18. This research was supported in part by a Helmsley Postdoctoral Fellowship at the Rockefeller University (to K.H.K.), funding from the Blavatnik Family Foundation (to E.N.), a research collaboration with the Waters Corporation (J.R.E.), and NIGMS grants R01GM052586 (to T.A.R.) and T32GM007753 (Harvard/MIT Medical Scientist Training Program). E.N. and T.A.R. are supported as Howard Hughes Medical Institute investigators. We thank P. Carvalho (Oxford University, UK) for providing reagents.
Author information
Authors and Affiliations
Contributions
N.O.B. purified proteins, solved the crystal structure, and performed biochemical experiments. K.H.K. performed cryo-EM data collection and analysis. N.O.B. and K.H.K. contributed equally and appear in alphabetical order in the author list. Z.J. purified and tested Cdc48 FFF mutants and performed biochemical experiments. T.E.W. performed hydrogen/deuterium-exchange experiments with supervision from J.R.E. V.S. performed cross-linking mass spectrometry analysis with supervision from E.N. T.W. oversaw the cryo-EM experiments. N.O.B. and T.A.R. wrote the manuscript. T.A.R. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Purification of proteins.
a, Gel-filtration chromatograms for C. thermophilum Cdc48 (blue), Ufd1/Npl4 (UN; green), and Cdc48/Ufd1/Npl4 (Cdc48/UN; red). The elution positions of molecular weight standards are shown in orange. b-d, Samples from the major peaks of the gel-filtration runs in a were subjected to SDS-PAGE and Coomassie blue staining. The single lane on the right in d shows the pooled peak fractions of the Cdc48 complex used for EM. M, molecular weight markers.
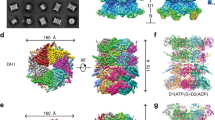
Supplementary Figure 2 Image processing of Cdc48 in the presence of ADP.
a, An area of a cryo-EM image of a vitrified sample is shown with some particles circled. Scale bar: 50 nm. b, Selected 2D class averages obtained with ISAC. Side length of individual averages: 32 nm. c, Initial 3D map obtained with VIPER. d, Image-processing workflow for 3D classification and refinement in RELION-1.4 that yielded density maps at resolutions of 8.9 Å (with no symmetry imposed) and 7.2 Å (with C6 symmetry imposed). See Methods section for details.
Supplementary Figure 3 Image processing of Cdc48 in the presence of ATPγS.
a, An area of a cryo-EM image of a vitrified sample is shown with some particles circled. Scale bar: 50 nm. b, Selected 2D class averages obtained with ISAC. Side length of individual averages: 25 nm. c, Initial 3D map obtained with VIPER. d, Image-processing workflow for 3D classification and refinement in RELION-1.4 that yielded density maps at resolutions of 10.3 Å (with no symmetry imposed) and 8.2 Å (with C6 symmetry imposed). See Methods section for details.
Supplementary Figure 4 FSC curves and local densities.
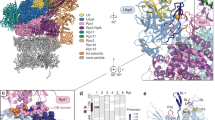
a, FSC curves calculated between independently refined half maps for the six-fold symmetrized density maps of Cdc48 alone in the presence of ADP (blue) or ATPγS (yellow), and for the density maps of the Cdc48/UN complex (no symmetry applied) in the presence of ADP (green) or ATPγS (red). b, Cross-validation FSC curves for the model and map of the Cdc48/UN complex with ATPγS showing no significant overfitting. c, Local resolution map of the Cdc48/cofactor complex structure obtained in the presence of ATPγS. Views are shown for a complete map (left) and a map after removing the front half (right). d, Representative cryo-EM densities with the fit models. ZF1, ZF2, Zn2+ fingers.
Supplementary Figure 5 Comparison of the Cdc48 and Cdc48–UN structures obtained in the presence of ADP and ATP-γS.
a, The cryo-EM density maps of Cdc48 obtained in the presence of ADP and ATPγS are shown separately (left and middle panels) as well as superimposed (right panel). All maps were low pass-filtered to 8 Å. b, As in a, but for the Cdc48/UN complex. All maps were low pass-filtered to 7 Å. c, The cryo-EM density map of the Cdc48/UN complexes obtained in the presence of ATPγS (left) showed density (blue) for the UBX-like domain of Npl4. An NMR structure of the complex of an N domain and the UBX-like domain (PDB: 2PJH) was docked into this cryo-EM density (inset on the right).
Supplementary Figure 6 Image processing of the Cdc48–UN complex in the presence of ADP.
a, An area of a cryo-EM image of a vitrified sample is shown with some particles circled. Scale bar: 50 nm. b, Selected 2D class averages obtained with RELION-1.4. Side length of individual averages: 33 nm. c, Initial 3D map obtained with RELION-1.4. d, Image-processing workflow for 3D classification and refinement in RELION-1.4 that yielded a density map at 6.7 Å resolution. Asterisks (*) indicate 3D classes that consist of particles with their N domains in the up-conformation. Daggers (†) indicate 3D classes that consist of particles with an extra density near an N domain that may attribute to the UBX-like domain of Npl4. See Methods section for details.
Supplementary Figure 7 Image processing of the Cdc48–UN complex in the presence of ATP-γS.
a, An area of a cryo-EM image of a vitrified sample is shown with some particles circled. Scale bar: 50 nm. b, Selected 2D class averages obtained with ISAC. Side length of individual averages: 33 nm. c, Initial 3D map obtained with cryoSPARC. d, Image-processing workflow for 3D classification and refinement in RELION-2 that yielded a density map at 4.3 Å. Asterisks (*) indicate 3D classes that consist of particles with their N domains in the up-conformation. Daggers (†) indicate 3D classes that consist of particles with an extra density near an N domain (only seen with a lower contouring threshold) that likely represents the UBX-like domain of Npl4. See Methods section for details.
Supplementary Figure 8 Analysis of interactions in the Cdc48–UN complex.
a, The C. thermophilum Cdc48/UN complex was treated with a bifunctional crosslinker and crosslinks were determined by mass spectrometry. See Suppl Data Set 2. b, Residues involved in crosslinks between Cdc48 and Npl4 are mapped onto the structure and shown as red and blue spheres, respectively. c, Purified C. thermophilum UN was treated with increasing concentrations of trypsin and subjected to SDS-PAGE and Coomassie blue staining. N, U: full-length Npl4 and Ufd1. F: a stable fragment of Npl4, identified by mass spectrometry as Npl4 129–602. d, Hydrogen/deuterium (H/D) exchange was performed with Npl4 residues 129–602 or with full-length Ufd1/Npl4. Regions of the Npl4 backbone protected by Ufd1 are colored in turquoise (residues 262–274, 316–327, 360–374, and 428–444) and mapped onto the structure of Npl4 in ribbon representation. The unassigned density from the cryo-EM map (in orange) is predicted to belong to Ufd1. e, Time courses showing relative deuterium exchange protection of the peptides covering regions highlighted in d (turquoise), as well as two peptides that were not protected by Ufd1 (222–237 and 527–545; yellow).
Supplementary Figure 9 Molecular-replacement model and omit map.
a, Two views of the EM density used as a molecular replacement model to solve the structure of the crystallized Npl4 fragment. The arrow indicates the extra density attributed to Ufd1, which does not reappear in the crystal structure. b, A simulated annealing composite omit map was calculated to assess model bias from the EM density map. The map is displayed at 1.0 σ with a carve of 1.8Å. Shown is the fit of the model into the density of the first Zn2+-finger domain. Arrows indicate residues whose side chains are not well represented in the map due to conformational flexibility. The red sphere indicates the zinc atom. c, As in b, but for the region of the second Zn2+ finger. d, As in b, but for the region of the β-strand finger. e, A second cofactor molecule cannot be fit above the ATPase ring, as there are significant clashes between the two Npl4 molecules (in blue and brown).
Supplementary Figure 10 Characterization of Cdc48 FFF mutants and Npl4 with mutations in the β-strand finger
. a, The indicated Cdc48 variants were subjected to gel filtration to analyze their oligomeric states. The position of hexamers is indicated. Note that mutation of the central Phe residue (FAF and AAA mutants) affects the migration of Cdc48 in gel filtration. b, Unfolding of poly-ubiquitinated Eos with wild-type Cdc48 and the indicated mutant of the conserved Tyr in the β-strand finger of Npl4. Data are shown as mean ± SD of n=3 technical replicates. c, A npl4-1 temperature-sensitive S. cerevisiae strain was transformed with a plasmid encoding wild-type Npl4 or the indicated mutations in insert-2, spotted in serial dilution, and incubated at the indicated temperatures for two days (30 and 37ºC) or three days (25ºC). d, Insert-2, in which mutations were made, is shown in purple and the Tyr in the β-strand finger is indicated (C. thermophilum Y472 corresponds to S. cerevisiae Y445).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10
Supplementary Dataset 1
Source data for unfolding assays
Rights and permissions
About this article
Cite this article
Bodnar, N.O., Kim, K.H., Ji, Z. et al. Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1–Npl4. Nat Struct Mol Biol 25, 616–622 (2018). https://doi.org/10.1038/s41594-018-0085-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0085-x
This article is cited by
-
The UBX domain in UBXD1 organizes ubiquitin binding at the C-terminus of the VCP/p97 AAA-ATPase
Nature Communications (2023)
-
The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions
Nature Structural & Molecular Biology (2023)
-
Mechanisms of substrate processing during ER-associated protein degradation
Nature Reviews Molecular Cell Biology (2023)
-
Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition
Nature Communications (2022)
-
Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state
Cell Discovery (2022)