Abstract
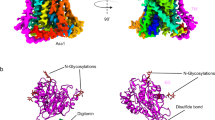
Human ASCT2 belongs to the SLC1 family of secondary transporters and is specific for the transport of small neutral amino acids. ASCT2 is upregulated in cancer cells and serves as the receptor for many retroviruses; hence, it has importance as a potential drug target. Here we used single-particle cryo-EM to determine a structure of the functional and unmodified human ASCT2 at 3.85-Å resolution. ASCT2 forms a homotrimeric complex in which each subunit contains a transport and a scaffold domain. Prominent extracellular extensions on the scaffold domain form the predicted docking site for retroviruses. Relative to structures of other SLC1 members, ASCT2 is in the most extreme inward-oriented state, with the transport domain largely detached from the central scaffold domain on the cytoplasmic side. This domain detachment may be required for substrate binding and release on the intracellular side of the membrane.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
25 June 2018
In the version of this article initially published, the links in the HTML to Supplementary Fig. 1 displayed an incorrect image for the figure. The errors have now been corrected.
References
Kanai, Y. et al. The SLC1 high-affinity glutamate and neutral amino acid transporter family. Mol. Aspects Med. 34, 108–120 (2013).
Slotboom, D. J., Konings, W. N. & Lolkema, J. S. Structural features of the glutamate transporter family. Microbiol. Mol. Biol. Rev. 63, 293–307 (1999).
Kanner, B. I. & Sharon, I. Active transport of L-glutamate by membrane vesicles isolated from rat brain. Biochemistry 17, 3949–3953 (1978).
Zerangue, N. & Kavanaugh, M. P. Flux coupling in a neuronal glutamate transporter. Nature 383, 634–637 (1996).
Danbolt, N. C. Glutamate uptake. Prog. Neurobiol. 65, 1–105 (2001).
Pochini, L., Scalise, M., Galluccio, M. & Indiveri, C. Membrane transporters for the special amino acid glutamine: structure/function relationships and relevance to human health. Front Chem. 2, 61 (2014).
Toda, K. et al. Clinical role of ASCT2 (SLC1A5) in KRAS-mutated colorectal cancer. Int. J. Mol. Sci. 18, E1632 (2017).
Kaira, K. et al. Relationship between CD147 and expression of amino acid transporters (LAT1 and ASCT2) in patients with pancreatic cancer. Am. J. Transl. Res. 7, 356–363 (2015).
Kim, S., Kim, D. H., Jung, W.-H. & Koo, J. S. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr. Relat. Cancer 20, 339–348 (2013).
Shimizu, K. et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br. J. Cancer 110, 2030–2039 (2014).
Wang, Q. et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 236, 278–289 (2015).
Bode, B. P., Kaminski, D. L., Souba, W. W. & Li, A. P. Glutamine transport in isolated human hepatocytes and transformed liver cells. Hepatology 21, 511–520 (1995).
Bröer, A., Rahimi, F. & Bröer, S. Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J. Biol. Chem. 291, 13194–13205 (2016).
Wang, Q. et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer 135, 1060–1071 (2014).
van Geldermalsen, M. et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35, 3201–3208 (2016).
Yoshikawa, R. et al. Simian retrovirus 4 induces lethal acute thrombocytopenia in Japanese macaques. J. Virol. 89, 3965–3975 (2015).
Shimode, S., Nakaoka, R., Shogen, H. & Miyazawa, T. Characterization of feline ASCT1 and ASCT2 as RD-114 virus receptor. J. Gen. Virol. 94, 1608–1612 (2013).
Tailor, C. S., Nouri, A., Zhao, Y., Takeuchi, Y. & Kabat, D. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 73, 4470–4474 (1999).
Lavillette, D. et al. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76, 6442–6452 (2002).
Yernool, D., Boudker, O., Jin, Y. & Gouaux, E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431, 811–818 (2004).
Boudker, O., Ryan, R. M., Yernool, D., Shimamoto, K. & Gouaux, E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature 445, 387–393 (2007).
Reyes, N., Ginter, C. & Boudker, O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature 462, 880–885 (2009).
Guskov, A., Jensen, S., Faustino, I., Marrink, S. J. & Slotboom, D. J. Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nat. Commun. 7, 13420 (2016).
Jensen, S., Guskov, A., Rempel, S., Hänelt, I. & Slotboom, D. J. Crystal structure of a substrate-free aspartate transporter. Nat. Struct. Mol. Biol. 20, 1224–1226 (2013).
Canul-Tec, J. C. et al. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544, 446–451 (2017).
Arkhipova, V., Guskov, A. & Slotboom, D. J. Analysis of the quality of crystallographic data and the limitations of structural models. J. Gen. Physiol. 149, 1091–1103 (2017).
Crisman, T. J., Qu, S., Kanner, B. I. & Forrest, L. R. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc. Natl Acad. Sci. USA 106, 20752–20757 (2009).
Pingitore, P. et al. Large scale production of the active human ASCT2 (SLC1A5) transporter in Pichia pastoris: functional and kinetic asymmetry revealed in proteoliposomes. Biochim. Biophys. Acta 1828, 2238–2246 (2013).
Scalise, M. et al. Transport mechanism and regulatory properties of the human amino acid transporter ASCT2 (SLC1A5). Amino Acids 46, 2463–2475 (2014).
Bröer, A., Wagner, C., Lang, F. & Bröer, S. Neutral amino acid transporter ASCT2 displays substrate-induced Na+ exchange and a substrate-gated anion conductance. Biochem. J. 346, 705–710 (2000).
Bröer, A. et al. The astroglial ASCT2 amino acid transporter as a mediator of glutamine efflux. J. Neurochem. 73, 2184–2194 (1999).
Utsunomiya-Tate, N., Endou, H. & Kanai, Y. Cloning and functional characterization of a system ASC-like Na+-dependent neutral amino acid transporter. J. Biol. Chem. 271, 14883–14890 (1996).
Zander, C. B., Albers, T. & Grewer, C. Voltage-dependent processes in the electroneutral amino acid exchanger ASCT2. J. Gen. Physiol. 141, 659–672 (2013).
Console, L., Scalise, M., Tarmakova, Z., Coe, I. R. & Indiveri, C. N-linked glycosylation of human SLC1A5 (ASCT2) transporter is critical for trafficking to membrane. Biochim. Biophys. Acta 1853, 1636–1645 (2015).
Marin, M., Lavillette, D., Kelly, S. M. & Kabat, D. N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J. Virol. 77, 2936–2945 (2003).
Bendahan, A., Armon, A., Madani, N., Kavanaugh, M. P. & Kanner, B. I. Arginine 447 plays a pivotal role in substrate interactions in a neuronal glutamate transporter. J. Biol. Chem. 275, 37436–37442 (2000).
Scopelliti, A. J., Font, J., Vandenberg, R. J., Boudker, O. & Ryan, R. M. Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5. Nat. Commun. 9, 38 (2018).
Akyuz, N. et al. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature 518, 68–73 (2015).
Wöhlert, D., Grötzinger, M. J., Kühlbrandt, W. & Yildiz, Ö. Mechanism of Na+-dependent citrate transport from the structure of an asymmetrical CitS dimer. eLife 4, e09375 (2015).
Coincon, M. et al. Crystal structures reveal the molecular basis of ion translocation in sodium/proton antiporters. Nat. Struct. Mol. Biol. 23, 248–255 (2016).
Drew, D. & Boudker, O. Shared molecular mechanisms of membrane transporters. Annu. Rev. Biochem. 85, 543–572 (2016).
Swier, L. J. Y. M., Guskov, A. & Slotboom, D. J. Structural insight in the toppling mechanism of an energy-coupling factor transporter. Nat. Commun. 7, 11072 (2016).
Geertsma, E. R., Nik Mahmood, N. A. B., Schuurman-Wolters, G. K. & Poolman, B. Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 (2008).
Biyani, N. et al. The interface between data collection and data processing in cryo-EM. J. Struct. Biol. 198, 124–133 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Bai, X.-C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. W. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Krissinel, E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319 (2015). W1.
Acknowledgements
We thank C. Indivieri (Università della Calabria, Italy) for kindly providing the P. pastoris expression strain, R.C. Prins for her help in preparation of cholesterol-containing proteoliposomes, M. Punter for help in setting up the image-processing cluster and B. Poolman for critical reading and discussion of the manuscript. This research was supported by NWO Vidi grant 723.014.002 to A.G.; NWO Veni grant 722.017.001 and Marie Skłodowska–Curie Individual Fellowship 749732 to C.P.; and NWO Vici grant 865.11.001 and European Research Council Starting Grant 282083 to D.J.S. C.G. thanks the SLAC National Accelerator Laboratory for financial support as part of the Panofsky fellowship program.
Author information
Authors and Affiliations
Contributions
A.G. conceived the project. Expression, purification and transport assays were performed by A.A.G. Initial cryo-EM experiments were carried out by C.G. Further and final cryo-EM sample preparation and data collection was done by A.A.G., G.T.O. and C.P. Cryo-EM image processing was carried out by A.A.G. and C.P. Model building and refinement were done by A.G. D.J.S. supervised the project at all stages and wrote the manuscript with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Cryo-EM reconstruction of ASCT2.
a,b, Representative cryo-EM image (a) and 2D-class averages (b) of vitrified ASCT2. c, Angular distribution plot of particles included in the final C3-symmetrized 3D reconstruction. The number of particles with the respective orientations is represented by length and colour of the cylinders (long and red: high number of particles; short and blue: low number of particles). d, Image processing work flow. e, Analysis of conformational heterogeneity in the dataset. 3D classification without any symmetry imposed where performed on the indicated particle dataset: after several rounds of 2D classification (I, on 628,015 particles) and after several rounds of 3D classification (II, on 307,619 particles). Particles obtained before particle polishing, which rendered a map of 3.9Å resolution were used to perform symmetry expansion followed by particle subtraction on the individual protomers, yielding a dataset of 552,240 particles (III). Resulting classes of the 3D classification without image alignment are shown. In all cases only the inward-facing state of ASCT2 was identified. f, Final reconstruction map coloured by local resolution as estimated by Relion. g, FSC plot of the final refined unmasked (grey) and masked (blue) map. The resolution at which the curve drops below the 0.143 threshold is indicated. A thumbnail of the mask used for FSC calculation overlaid on the atomic model is shown in the upper right corner. h, FSC curves of the refined model versus the map of ASCT2 for cross-validation. The purple shows the FSC curves for the refined model compared to the full masked dataset (FSCsum); light grey, FSC curve for the refined model compared to the masked half-map 1 (FSCwork, used during validation refinement); dark grey, refined model compared to the masked half- map 2 (FSCfree, not used during validation refinement). Dashed lines, FSC threshold used for FSCsum of 0.5 and for FSCfree/work of 0.143.
Supplementary Figure 2 Cryo-EM density.
Sections of the cryo-EM density of the ASCT2 map superimposed on the respective refined models. Models are shown as sticks and structural elements are labelled. Transmembrane helices of the transport domain are coloured in blue, of the scaffold domain in yellow and the loop between TM4b and TM4c is shown in red. Densities sharpened with a b-factor of −225 Å2 were plotted at 5σ, except for the loop between TM4b-TM4c, which was contoured at 3σ.
Supplementary Figure 3 Superposition and orientation of domains in ASCT2 and EAAT1.
Superposition of transport (a) and scaffold (b) domains of ASCT2 (bright blue and bright yellow) with EAAT1 (light blue and light orange, PDB 5LLU), respectively. Rmsds are ~ 1Å. c,d, The scaffold domains of ASCT2 (yellow ribbon) and EAAT1 (orange ribbon, PDB 5LLU) were aligned structurally, revealing large differences in the position of the transport domains (dark and light blue surfaces, respectively) c, The movement form the outward (as observed in EAAT1) to the inward (as seen in ASCT2) state is indicated with an arrow. d, A pair of equivalent residues (Glu444 of ASCT2 and T456 of EAAT1) is highlighted in red to emphasize the movement of ~ 25 Å.
Supplementary Figure 4 Visualization of the domain interface and putative lipid density.
A single protomer of ASCT2 (a), EAAT1 (PDB 5LLU) (b), GltPh in the locked state (PDB 4X2S, chain A) (c) and GltPh in the unlocked state (PDB 4X2S, chain C) (d) viewed from the membrane plane. The scaffold domain is depicted as yellow ribbon and the transport domain in grey surface representation) with the interacting parts in black. The surface area of the interfaces are extensive in EAAT1 and locked GltPh. The transport domains in ACST2 and unlocked GltPh are more detached from the scaffold domain, but the shapes of their interaction surfaces are different, with most of the interacting residues in ASCT2 located in a narrow strip on the extracellular side. e, Patches of densities (green mesh at 4σ) observed between scaffold (yellow) and transport domain (blue) can accommodate cholesteryl hemisuccinate molecules shown as pink sticks, but the identities of the molecules could not be assigned unambiguously.
Supplementary Figure 5 Pseudosymmetrical relation between the transport domains of EAAT1 and ASCT2.
Transport domain of the outward-facing structure of EAAT1 (PDB 5LLU, pale colors, panel a, d), and the inward-facing structure of ASCT2 (bright colors, panel c, f) were structurally aligned on their scaffold domains (not visible) and are depicted from two perpendicular viewpoints in the plane of the membrane. b,e: The pseudo-symmetrical organization observed between transport domains in both structures becomes apparent when they are overlaid. TM3 (bright blue) is pseudo-symmetrical to TM6 (pale blue), TM6 (bright pink) to TM3 (pale pink) and HP1 (bright yellow and orange) to HP2 (pale yellow and pale orange). In the top row the pseudo two-fold axis is indicated as a dashed arrow. In the bottom row the two-fold axis is indicated as a black dot and points towards the reader.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5
Supplementary Note
Sequence alignment. Sequence alignment of human ASCT2 with all other human SLC1 family members and archaeal homologues GltPh and GltTk. Transmembrane segments are indicated with numbered cylinders and colored yellow and blue for scaffold and transport domains, respectively. The protruding loop forming the ‘antenna’ is shown with the red dashed box. The sequence conservation is shown with different shades of violet. The residues forming a putative salt bridge between the domains in ASCT2 and the binding-site residues are indicated by grey squares and triangles, respectively. The transport domain of ASCT2 shares 50% and 34% identical residues with those of EAAT1 and GltPh, respectively. The scaffold domains 42% and 27%, respectively.
Supplementary Video 1
ASCT2 structure. The cryo-EM density (sharpened with a B-factor of -225 Å2) of the human inward-facing ASCT2 amino acid exchanger, is shown with the modelled structure superimposed. For clarity only one protomer of the trimer is shown, the transport domain is coloured in blue, the scaffold domain in yellow and the extracellular loop between TM4b and TM4c is shown in red. The view is from within the membrane.
Supplementary Video 2
Putative conformational changes of the transport domain in human SLC1. Morph between the inward-facing human ASCT2 structure and the outward-facing human EAAT1 (PDB-ID: 5llm) structure. The structures were superimposed on their scaffold domains. For clarity only one protomer of the homotrimer is shown. The scaffold domain is coloured in yellow and the transport domain in blue with the hairpins HP1 depicted in light purple and HP2 in purple. The hairpins of the inward-facing unlocked GltPh (PDB-ID: 4X2S, chain C) structure are shown in green. The membrane boundaries are indicated by grey lines. Note the accessibility of HP2 to the cytoplasm.
Rights and permissions
About this article
Cite this article
Garaeva, A.A., Oostergetel, G.T., Gati, C. et al. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat Struct Mol Biol 25, 515–521 (2018). https://doi.org/10.1038/s41594-018-0076-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0076-y
This article is cited by
-
The NERP-4–SNAT2 axis regulates pancreatic β-cell maintenance and function
Nature Communications (2023)
-
TFEB inhibition induces melanoma shut-down by blocking the cell cycle and rewiring metabolism
Cell Death & Disease (2023)
-
Structural basis of ligand binding modes of human EAAT2
Nature Communications (2022)
-
Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2
Nature Communications (2022)
-
Molecular Basis of Coupled Transport and Anion Conduction in Excitatory Amino Acid Transporters
Neurochemical Research (2022)