Abstract
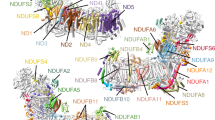
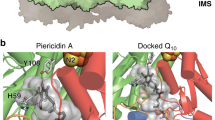
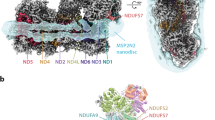
Complex I (NADH:ubiquinone oxidoreductase) uses the reducing potential of NADH to drive protons across the energy-transducing inner membrane and power oxidative phosphorylation in mammalian mitochondria. Recent cryo-EM analyses have produced near-complete models of all 45 subunits in the bovine, ovine and porcine complexes and have identified two states relevant to complex I in ischemia–reperfusion injury. Here, we describe the 3.3-Å structure of complex I from mouse heart mitochondria, a biomedically relevant model system, in the ‘active’ state. We reveal a nucleotide bound in subunit NDUFA10, a nucleoside kinase homolog, and define mechanistically critical elements in the mammalian enzyme. By comparisons with a 3.9-Å structure of the ‘deactive’ state and with known bacterial structures, we identify differences in helical geometry in the membrane domain that occur upon activation or that alter the positions of catalytically important charged residues. Our results demonstrate the capability of cryo-EM analyses to challenge and develop mechanistic models for mammalian complex I.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hirst, J. Mitochondrial complex I. Annu. Rev. Biochem. 82, 551–575 (2013).
Fassone, E. & Rahman, S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J. Med. Genet. 49, 578–590 (2012).
Hirst, J., Carroll, J., Fearnley, I. M., Shannon, R. J. & Walker, J. E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604, 135–150 (2003).
Stroud, D. A. et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126 (2016).
Sazanov, L. A. & Hinchliffe, P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 (2006).
Efremov, R. G. & Sazanov, L. A. Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 (2011).
Baradaran, R., Berrisford, J. M., Minhas, G. S. & Sazanov, L. A. Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013).
Vinothkumar, K. R., Zhu, J. & Hirst, J. Architecture of mammalian respiratory complex I. Nature 515, 80–84 (2014).
Zhu, J. et al. Structure of subcomplex Iβ of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc. Natl. Acad. Sci. USA 112, 12087–12092 (2015).
Zhu, J., Vinothkumar, K. R. & Hirst, J. Structure of mammalian respiratory complex I. Nature 536, 354–358 (2016).
Fiedorczuk, K. et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature 538, 406–410 (2016).
Wu, M., Gu, J., Guo, R., Huang, Y. & Yang, M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609 (2016).
Guo, R., Zong, S., Wu, M., Gu, J. & Yang, M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170, 1247–1257 (2017).
Zickermann, V. et al. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347, 44–49 (2015).
Cooley, R. B., Arp, D. J. & Karplus, P. A. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J. Mol. Biol. 404, 232–246 (2010).
Kumar, P. & Bansal, M. Dissecting π-helices: sequence, structure and function. FEBS J. 282, 4415–4432 (2015).
Kaila, V. R. I., Wikström, M. & Hummer, G. Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I. Proc. Natl. Acad. Sci. USA 111, 6988–6993 (2014).
Sharma, V. et al. Redox-induced activation of the proton pump in the respiratory complex I. Proc. Natl. Acad. Sci. USA 112, 11571–11576 (2015).
Di Luca, A., Gamiz-Hernandez, A. P. & Kaila, V. R. I. Symmetry-related proton transfer pathways in respiratory complex I. Proc. Natl. Acad. Sci. USA 114, E6314–E6321 (2017).
Haapanen, O. & Sharma, V. Role of water and protein dynamics in proton pumping by respiratory complex I. Sci. Rep. 7, 7747 (2017).
Blaza, J. N., Vinothkumar, K. R. & Hirst, J. Structure of the deactive state of mammalian respiratory complex I. Structure 26, 312–319 (2018).
Kotlyar, A. B. & Vinogradov, A. D. Slow active/inactive transition of the mitochondrial NADH-ubiquinone reductase. Biochim. Biophys. Acta 1019, 151–158 (1990).
Vinogradov, A. D. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim. Biophys. Acta 1364, 169–185 (1998).
Babot, M., Birch, A., Labarbuta, P. & Galkin, A. Characterisation of the active/de-active transition of mitochondrial complex I. Biochim. Biophys. Acta 1837, 1083–1092 (2014).
Galkin, A. & Moncada, S. Modulation of the conformational state of mitochondrial complex I as a target for therapeutic intervention. Interface Focus 7, 20160104 (2017).
Galkin, A., Abramov, A. Y., Frakich, N., Duchen, M. R. & Moncada, S. Lack of oxygen deactivates mitochondrial complex I: implications for ischemic injury? J. Biol. Chem. 284, 36055–36061 (2009).
Chouchani, E. T. et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 19, 753–759 (2013).
Russo, C. J. & Passmore, L. A. Electron microscopy: ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014).
Meyerson, J. R. et al. Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Sci. Rep. 4, 7084 (2014).
Letts, J. A., Fiedorczuk, K. & Sazanov, L. A. The architecture of respiratory supercomplexes. Nature 537, 644–648 (2016).
Di Luca, A. & Kaila, V. R. I. Global collective motions in the mammalian and bacterial respiratory complex I. Biochim. Biophys. Acta 1859, 326–332 (2018).
Hunte, C. Specific protein-lipid interactions in membrane proteins. Biochem. Soc. Trans. 33, 938–942 (2005).
Sharpley, M. S., Shannon, R. J., Draghi, F. & Hirst, J. Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry 45, 241–248 (2006).
Eriksson, S., Munch-Petersen, B., Johansson, K. & Eklund, H. Structure and function of cellular deoxyribonucleoside kinases. Cell. Mol. Life Sci. 59, 1327–1346 (2002).
Sabini, E., Hazra, S., Ort, S., Konrad, M. & Lavie, A. Structural basis for substrate promiscuity of dCK. J. Mol. Biol. 378, 607–621 (2008).
Mikkelsen, N. E. et al. Structural basis for feedback inhibition of the deoxyribonucleoside salvage pathway: studies of the Drosophila deoxyribonucleoside kinase. Biochemistry 42, 5706–5712 (2003).
Schilling, B. et al. Mass spectrometric identification of a novel phosphorylation site in subunit NDUFA10 of bovine mitochondrial complex I. FEBS Lett. 579, 2485–2490 (2005).
Elurbe, D. M. & Huynen, M. A. The origin of the supernumerary subunits and assembly factors of complex I: a treasure trove of pathway evolution. Biochim. Biophys. Acta 1857, 971–979 (2016).
Hirst, J. & Roessler, M. M. Energy conversion, redox catalysis and generation of reactive oxygen species by respiratory complex I. Biochim. Biophys. Acta 1857, 872–883 (2016).
Carroll, J., Ding, S., Fearnley, I. M. & Walker, J. E. Post-translational modifications near the quinone binding site of mammalian complex I. J. Biol. Chem. 288, 24799–24808 (2013).
Zwicker, K. et al. The redox-Bohr group associated with iron-sulfur cluster N2 of complex I. J. Biol. Chem. 281, 23013–23017 (2006).
Le Breton, N. et al. Using hyperfine electron paramagnetic resonance spectroscopy to define the proton-coupled electron transfer reaction at Fe-S cluster N2 in respiratory complex I. J. Am. Chem. Soc. 139, 16319–16326 (2017).
Gamiz-Hernandez, A. P., Jussupow, A., Johansson, M. P. & Kaila, V. R. I. Terminal electron−proton transfer dynamics in the quinone reduction of respiratory complex I. J. Am. Chem. Soc. 139, 16282–16288 (2017).
Hryc, C. F. et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc. Natl. Acad. Sci. USA 114, 3103–3108 (2017).
Fedor, J. G., Jones, A. J. Y., Di Luca, A., Kaila, V. R. I. & Hirst, J. Correlating kinetic and structural data on ubiquinone binding and reduction by respiratory complex I. Proc. Natl. Acad. Sci. USA 114, 12737–12742 (2017).
Birrell, J. A. & Hirst, J. Truncation of subunit ND2 disrupts the threefold symmetry of the antiporter-like subunits in complex I from higher metazoans. FEBS Lett. 584, 4247–4252 (2010).
Belevich, G., Knuuti, J., Verkhovsky, M. I., Wikström, M. & Verkhovskaya, M. Probing the mechanistic role of the long α-helix in subunit L of respiratory complex I from Escherichia coli by site-directed mutagenesis. Mol. Microbiol. 82, 1086–1095 (2011).
Zhu, S. & Vik, S. B. Constraining the lateral helix of respiratory complex I by cross-linking does not impair enzyme activity or proton translocation. J. Biol. Chem. 290, 20761–20773 (2015).
Steimle, S. et al. Asp563 of the horizontal helix of subunit NuoL is involved in proton translocation by the respiratory complex I. FEBS Lett. 586, 699–704 (2012).
Sazinsky, M. H. & Lippard, S. J. Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the α-subunit. J. Am. Chem. Soc. 127, 5814–5825 (2005).
Calvaruso, M. A. et al. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 21, 115–120 (2012).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017).
Chovancova, E. et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 8, e1002708 (2012).
Fernández-Vizarra, E. et al. Isolation of mitochondria for biogenetical studies: an update. Mitochondrion 10, 253–262 (2010).
Carroll, J., Altman, M. C., Fearnley, I. M. & Walker, J. E. Identification of membrane proteins by tandem mass spectrometry of protein ions. Proc. Natl. Acad. Sci. USA 104, 14330–14335 (2007).
Bridges, H. R., Mohammed, K., Harbour, M. E. & Hirst, J. Subunit NDUFV3 is present in two distinct isoforms in mammalian complex I. Biochim. Biophys. Acta 1858, 197–207 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Heymann, J. B. & Belnap, D. M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Touw, W. G. et al. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 43, D364–D368 (2015).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank the staff at the Astbury Biostructure Laboratory, University of Leeds (funded by the University of Leeds and Wellcome Trust 108466/Z/15/Z) and at the UK National Electron Bio-Imaging Centre (eBIC) at the Diamond Light Source for assistance with cryo-EM data collection, the staff at the Phenomics Animal Care Facility, S. Ding and I. Fearnley (Cambridge) for mass spectrometry analyses, and M. Hartley and A. Raine (Cambridge) for IT support. This work was supported by Medical Research Council grant numbers MC_U105663141 (J.H.), MC_UU_00015/2 (J.H.) and MC_UU_00015/5 (C.V.).
Author information
Authors and Affiliations
Contributions
A.-N.A.A. prepared and characterized mouse complex I assisted by H.R.B. and C.V.; J.N.B. prepared cryo-EM grids and collected data, assisted by S.R. and S.P.M.; A.-N.A.A. and J.N.B. processed cryo-EM data assisted by H.R.B., S.R. and S.P.M.; J.N.B. built the models assisted by A.-N.A.A.; J.H. analyzed and interpreted the models assisted by A.-N.A.A., J.N.B. and H.R.B; J.H. directed the project and wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Resolution estimates of the maps for the active and deactive states of complex I.
a, b: data on the active complex. c, d: data on the deactive complex. The estimated resolutions, defined where the line at FSC = 0.143 crosses the red curve, are 3.3 Å for the active complex (a) and 3.9 Å for the deactive complex (c). In both cases the refined models agree well with the maps, as shown by the map vs. model FSC curves (blue). In b) and d), local resolutions were estimated using the Local Resolution function in RELION with default parameters and plotted using UCSF Chimera.
Supplementary Figure 2
Classification and refinement of the cryo-EM density maps for the active and deactive states of mouse complex I.
Supplementary Figure 3
Example densities (carved at 2 Å radius, threshold level 0.065) drawn from different parts of the map of active mouse complex I.
Supplementary Figure 4
Example densities (carved at 2 Å radius, threshold level 0.04) of phosholipids from the map of active mouse complex I.
Supplementary Figure 5 Mass spectrometry evidence for the phosphorylation of bovine NDUFA10-Ser36.
a) Intact mass measurements revealed two masses for NDUFA10, for the unmodified and singly phosphorylated versions (present at the point of measurement). b) Spectrum of fragment ions produced by ETD of a doubly charged precursor ion 942.48 (m/z) with the precursor ion truncated to 10% relative intensity. The observed neutral peptide mass for the phosphorylated peptide is 1882.955 Da, relative to a calculated mass of 1882.959 Da. The observed peptides are mapped onto the amino acid sequence where the phosphorylation site is marked and c is carbamidomethylcysteine. The z7 and z8 fragments confirm Ser36, rather than Ser33, as the site of phosphorylation. No additional phosphorylated peptides from NDUFA10 were observed.
Supplementary Figure 6
Densities and models for TMH8 in subunits ND2, ND4 and ND5 of mouse complex I (carved at 2 Å radius, threshold level 0.065).
Supplementary information
Rights and permissions
About this article
Cite this article
Agip, AN.A., Blaza, J.N., Bridges, H.R. et al. Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat Struct Mol Biol 25, 548–556 (2018). https://doi.org/10.1038/s41594-018-0073-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0073-1
This article is cited by
-
N1-methylation of adenosine (m1A) in ND5 mRNA leads to complex I dysfunction in Alzheimer’s disease
Molecular Psychiatry (2024)
-
SCAF1 drives the compositional diversity of mammalian respirasomes
Nature Structural & Molecular Biology (2024)
-
Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4−/− mouse
The EMBO Journal (2024)
-
A lethal mitonuclear incompatibility in complex I of natural hybrids
Nature (2024)
-
Mechanism of rotenone binding to respiratory complex I depends on ligand flexibility
Scientific Reports (2023)