Abstract
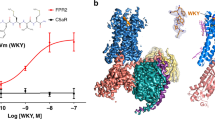
Platelet-activating-factor receptor (PAFR) responds to platelet-activating factor (PAF), a phospholipid mediator of cell-to-cell communication that exhibits diverse physiological effects. PAFR is considered an important drug target for treating asthma, inflammation and cardiovascular diseases. Here we report crystal structures of human PAFR in complex with the antagonist SR 27417 and the inverse agonist ABT-491 at 2.8-Å and 2.9-Å resolution, respectively. The structures, supported by molecular docking of PAF, provide insights into the signal-recognition mechanisms of PAFR. The PAFR–SR 27417 structure reveals an unusual conformation showing that the intracellular tips of helices II and IV shift outward by 13 Å and 4 Å, respectively, and helix VIII adopts an inward conformation. The PAFR structures, combined with single-molecule FRET and cell-based functional assays, suggest that the conformational change in the helical bundle is ligand dependent and plays a critical role in PAFR activation, thus greatly extending knowledge about signaling by G-protein-coupled receptors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
08 May 2019
In the version of this article initially published, Supplementary Table 1 and the Supplementary Note were omitted from the Supplementary Text and Figures file. The error has been corrected.
References
Hwang, S. B. Specific receptors of platelet-activating factor, receptor heterogeneity, and signal transduction mechanisms. J. Lipid Mediat. 2, 123–158 (1990).
Braquet, P., Touqui, L., Shen, T. Y. & Vargaftig, B. B. Perspectives in platelet-activating factor research. Pharmacol. Rev. 39, 97–145 (1987).
Chao, W. & Olson, M. S. Platelet-activating factor: receptors and signal transduction. Biochem. J. 292, 617–629 (1993).
Dupré, D. J., Le Gouill, C., Rola-Pleszczynski, M. & Stanková, J. Inverse agonist activity of selected ligands of platelet-activating factor receptor. J. Pharmacol. Exp. Ther. 299, 358–365 (2001).
Herbert, J. M. et al. Biochemical and pharmacological activities of SR 27417, a highly potent, long-acting platelet-activating factor receptor antagonist. J. Pharmacol. Exp. Ther. 259, 44–51 (1991).
Albert, D. H. et al. Pharmacology of ABT-491, a highly potent platelet-activating factor receptor antagonist. Eur. J. Pharmacol. 325, 69–80 (1997).
Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure 20, 967–976 (2012).
Thorsen, T. S., Matt, R., Weis, W. I. & Kobilka, B. K. Modified T4 lysozyme fusion proteins facilitate G protein-coupled receptor crystallogenesis. Structure 22, 1657–1664 (2014).
Ballesteros, J. & Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 (1995).
Zhang, K. et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509, 115–118 (2014).
Zhang, J. et al. Agonist-bound structure of the human P2Y12 receptor. Nature 509, 119–122 (2014).
Hanson, M. A. et al. Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 (2012).
Chrencik, J. E. et al. Crystal structure of antagonist bound human lysophosphatidic acid receptor 1. Cell 161, 1633–1643 (2015).
Srivastava, A. et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124–127 (2014).
Hua, T. et al. Crystal structure of the human cannabinoid receptor CB1. Cell 167, 750–762.e14 (2016).
Curtin, M. L. et al. Discovery and evaluation of a series of 3-acylindole imidazopyridine platelet-activating factor antagonists. J. Med. Chem. 41, 74–95 (1998).
Ryan, S. D., Harris, C. S., Carswell, C. L., Baenziger, J. E. & Bennett, S. A. Heterogeneity in the sn-1 carbon chain of platelet-activating factor glycerophospholipids determines pro- or anti-apoptotic signaling in primary neurons. J. Lipid Res. 49, 2250–2258 (2008).
Wess, J., Han, S. J., Kim, S. K., Jacobson, K. A. & Li, J. H. Conformational changes involved in G-protein-coupled-receptor activation. Trends Pharmacol. Sci. 29, 616–625 (2008).
Kuwasako, K., Kitamura, K., Nagata, S., Hikosaka, T. & Kato, J. Structure-function analysis of helix 8 of human calcitonin receptor-like receptor within the adrenomedullin 1 receptor. Peptides 32, 144–149 (2011).
Wacker, D., Stevens, R. C. & Roth, B. L. How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017).
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011).
Park, J. H., Scheerer, P., Hofmann, K. P., Choe, H. W. & Ernst, O. P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 (2008).
Xu, F. et al. Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 (2011).
Zhang, D. et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 520, 317–321 (2015).
Ernst, O. P. et al. Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin alpha and gamma subunits. J. Biol. Chem. 275, 1937–1943 (2000).
Delos Santos, N. M., Gardner, L. A., White, S. W. & Bahouth, S. W. Characterization of the residues in helix 8 of the human β1-adrenergic receptor that are involved in coupling the receptor to G proteins. J. Biol. Chem. 281, 12896–12907 (2006).
Sounier, R. et al. Propagation of conformational changes during μ-opioid receptor activation. Nature 524, 375–378 (2015).
Liang, Y. L. et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123 (2017).
Zhang, Y. et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253 (2017).
Liang, Y. L. et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555, 121–125 (2018).
Zhang, H. et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature 544, 327–332 (2017).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68, 368–380 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Vanommeslaeghe, K. & MacKerell, A. D. Jr. Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Vanommeslaeghe, K., Raman, E. P. & MacKerell, A. D. Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Miyamoto, S. & Kollman, P. A. Settle: an analytical version of the shake and rattle algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Chatterjee, A., Sun, S. B., Furman, J. L., Xiao, H. & Schultz, P. G. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837 (2013).
Schmied, W. H., Elsässer, S. J., Uttamapinant, C. & Chin, J. W. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 136, 15577–15583 (2014).
Juette, M. F. et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016).
Acknowledgements
This work was supported by the National Key R&D Program of China, 2018YFA0507000; CAS Strategic Priority Research Program grant XDB08020000 (B.W., X.C.Z., Z.R. and X.L.); the Key Research Program of Frontier Sciences, CAS, grants QYZDB-SSW-SMC024 (B.W.) and QYZDB-SSW-SMC054 (Q.Z.); the National Science Foundation of China, grants 81525024 (Q.Z.) and 31301163 (C.X.); and the Program of Introducing Talents of Discipline to the Universities of the Ministry of Education (grant B08029) (J.L.). The authors thank M. Hanson, V. Cherezov and V. Katritch for careful review and scientific feedback on the manuscript; H. Zhang for guidance in handling radiolabeled chemicals; and J. W. Chin (Medical Research Council Laboratory of Molecular Biology, Cambridge) for providing plasmids (U6-PylT*)4/EF1α-PylRS, (U6-PylT*)4/EF1α-sfGFP(TAG) and peRF1 (E55D). The synchrotron radiation experiments were performed at BL41XU of SPring-8 with approval from the Japan Synchrotron Radiation Research Institute (proposal nos. 2014B1057, 2015A1026, 2015A1027, 2015B2026 and 2015B2027). We thank the BL41XU beamline staff members K. Hasegawa, H. Okumura and H. Murakami for help with X-ray data collection.
Author information
Authors and Affiliations
Contributions
C.C. and Q.T. optimized the construct; developed the purification procedure and purified the PAFR proteins for crystallization; and performed crystallization trials and optimized crystallization conditions. C.X. and Yiwei Zhou designed, performed and analyzed Ca2+-flux and IP-accumulation assays of WT and mutant PAFRs. L.H. and C.C. designed, performed and analyzed smFRET assays. L.Y. performed and analyzed MD simulations and docking assays. C.C. and Ye Zhou designed, performed and analyzed ligand-binding assays of WT and mutant PAFRs. A.Q. and M.L. assisted in construct and crystal optimization. C.Y. expressed the PAFR proteins. G.W.H. assisted in structure refinement. X.W. and X.L. helped to develop the initial expression and purification protocol for PAFR. H.Y. oversaw computational assays. Z.R. oversaw structure analysis and interpretation. H.J. oversaw computational assays and structure analysis and interpretation. Y. Zhao oversaw smFRET assays. J.L. oversaw Ca2+-flux and IP-accumulation assays, and edited the manuscript. R.C.S. assisted in structure analysis and interpretation, and edited the manuscript. Q.Z. oversaw construct design, collected crystal diffraction data, solved the PAFR structures and assisted with manuscript preparation. X.C.Z. and B.W. initiated the project, planned and analyzed experiments, solved the structures, supervised the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Table 1 and Supplementary Note
Rights and permissions
About this article
Cite this article
Cao, C., Tan, Q., Xu, C. et al. Structural basis for signal recognition and transduction by platelet-activating-factor receptor. Nat Struct Mol Biol 25, 488–495 (2018). https://doi.org/10.1038/s41594-018-0068-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-018-0068-y
This article is cited by
-
Bitter taste receptor activation by cholesterol and an intracellular tastant
Nature (2024)
-
PAFAH2 suppresses synchronized ferroptosis to ameliorate acute kidney injury
Nature Chemical Biology (2024)
-
Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding
Nature Communications (2023)
-
Insights into divalent cation regulation and G13-coupling of orphan receptor GPR35
Cell Discovery (2022)
-
Universal platform for the generation of thermostabilized GPCRs that crystallize in LCP
Nature Protocols (2022)