Abstract
The development of physical dependence and addiction disorders due to misuse of opioid analgesics is a major concern with pain therapeutics. We developed a mouse model of oxycodone exposure and subsequent withdrawal in the presence or absence of chronic neuropathic pain. Oxycodone withdrawal alone triggered robust gene expression adaptations in the nucleus accumbens, medial prefrontal cortex and ventral tegmental area, with numerous genes and pathways selectively affected by oxycodone withdrawal in mice with peripheral nerve injury. Pathway analysis predicted that histone deacetylase (HDAC) 1 is a top upstream regulator in opioid withdrawal in nucleus accumbens and medial prefrontal cortex. The novel HDAC1/HDAC2 inhibitor, Regenacy Brain Class I HDAC Inhibitor (RBC1HI), attenuated behavioral manifestations of oxycodone withdrawal, especially in mice with neuropathic pain. These findings suggest that inhibition of HDAC1/HDAC2 may provide an avenue for patients with chronic pain who are dependent on opioids to transition to non-opioid analgesics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq data are available on the NCBI Gene Expression Omnibus under accession GSE223541. Other data from this study can be made available upon request.
Change history
22 January 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41593-024-01579-6
References
Darcq, E. & Kieffer, B. L. Opioid receptors: drivers to addiction? Nat. Rev. Neurosci. 19, 499–514 (2018).
Serafini, R. A., Pryce, K. D. & Zachariou, V. The mesolimbic dopamine system in chronic pain and associated affective comorbidities. Biol. Psychiatry 87, 64–73 (2020).
Finnerup, N. B. et al. Pharmacotherapy for neuropathic pain in adults: a systematic review and meta-analysis. Lancet Neurol. 14, 162–173 (2015).
Dworkin, R. H. et al. Pharmacologic management of neuropathic pain: evidence-based recommendations. Pain 132, 237–251 (2007).
Cruccu, G. Treatment of painful neuropathy. Curr. Opin. Neurol. 20, 531–535 (2007).
Hooten, W. M., Lamer, T. J. & Twyner, C. Opioid-induced hyperalgesia in community-dwelling adults with chronic pain. Pain 156, 1145–1152 (2015).
Volkow, N. D., Michaelides, M. & Baler, R. The neuroscience of drug reward and addiction. Physiol. Rev. 99, 2115–2140 (2019).
Zhang, Y. et al. Chronic oxycodone self-administration altered reward-related genes in the ventral and dorsal striatum of C57BL/6J mice: an RNA-seq analysis. Neuroscience 393, 333–349 (2018).
Koob, G. F. & Volkow, N. D. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry 3, 760–773 (2016).
Baliki, M. N., Chang, P. C., Baria, A. T., Centeno, M. V. & Apkarian, A. V. Resting-sate functional reorganization of the rat limbic system following neuropathic injury. Sci. Rep. 4, 6186 (2014).
Apkarian, V. A., Hashmi, J. A. & Baliki, M. N. Pain and the brain: specificity and plasticity of the brain in clinical chronic pain. Pain 152, S49–S64 (2011).
Seminowicz, D. A. et al. Pain-related nucleus accumbens function: modulation by reward and sleep disruption. Pain 160, 1196–1207 (2019).
Schwartz, N. et al. Chronic pain. Decreased motivation during chronic pain requires long-term depression in the nucleus accumbens. Science 345, 535–542 (2014).
Browne, C. J., Godino, A., Salery, M. & Nestler, E. J. Epigenetic mechanisms of opioid addiction. Biol. Psychiatry 87, 22–33 (2020).
Jullie, D., Gondin, A. B., von Zastrow, M. & Canals, M. Opioid pharmacology under the microscope. Mol. Pharm. 98, 425–432 (2020).
Baliki, M. N., Geha, P. Y., Fields, H. L. & Apkarian, A. V. Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron 66, 149–160 (2010).
Darnall, B. To treat pain, study people in all their complexity. Nature 557, 7 (2018).
McPherson, M. L. et al. Safe and appropriate use of methadone in hospice and palliative care: expert consensus white paper. J. Pain Symptom Manag. 57, 635–645 (2019).
Bertin, C. et al. Analgesic opioid use disorders in patients with chronic non-cancer pain: a holistic approach for tailored management. Neurosci. Biobehav. Rev. 121, 160–174 (2021).
Delorme, J. et al. Prevalence and characteristics of chronic pain in buprenorphine and methadone-maintained patients. Front. Psychiatry 12, 641430 (2021).
Descalzi, G. et al. Neuropathic pain promotes adaptive changes in gene expression in brain networks involved in stress and depression. Sci. Signal. 10, eaaj1549 (2017).
Mitsi, V. et al. RGS9-2–controlled adaptations in the striatum determine the onset of action and efficacy of antidepressants in neuropathic pain states. Proc. Natl Acad. Sci. USA 112, E5088–E5097 (2015).
Mitsi, V. & Zachariou, V. Modulation of pain, nociception, and analgesia by the brain reward center. Neuroscience 338, 81–92 (2016).
Corder, G. et al. An amygdalar neural ensemble that encodes the unpleasantness of pain. Science 363, 276–281 (2019).
Ren, W. et al. Adaptive alterations in the mesoaccumbal network after peripheral nerve injury. Pain 162, 895–906 (2021).
Shields, S. D., Eckert, W. A. 3rd & Basbaum, A. I. Spared nerve injury model of neuropathic pain in the mouse: a behavioral and anatomic analysis. J. Pain 4, 465–470 (2003).
Avrampou, K. et al. RGS4 maintains chronic pain symptoms in rodent models. J. Neurosci. 39, 8291–8304 (2019).
Anderson, E. M. et al. Knockdown of the histone di-methyltransferase G9a in nucleus accumbens shell decreases cocaine self-administration, stress-induced reinstatement, and anxiety. Neuropsychopharmacology 44, 1370–1376 (2019).
Cobos, E. J. et al. Inflammation-induced decrease in voluntary wheel running in mice: a nonreflexive test for evaluating inflammatory pain and analgesia. Pain 153, 876–884 (2012).
Lee, E. -H., Park, J. -Y., Lee, Y. & Han, P. -L. Sociability and social novelty preference tests using a U-shaped two-choice field. Bio. Protoc. 8, e2853 (2018).
Lustberg, D. et al. Noradrenergic circuits in the forebrain control affective responses to novelty. Psychopharmacol. 237, 3337–3355 (2020).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Avey, D. et al. Single-cell RNA-seq uncovers a robust transcriptional response to morphine by glia. Cell Rep. 24, 3619–3629 (2018).
Ray, M. H., Williams, B. R., Kuppe, M. K., Bryant, C. D. & Logan, R. W. A glitch in the matrix: the role of extracellular matrix remodeling in opioid use disorder. Front. Integr. Neurosci. 16, 899637 (2022).
Flaisher-Grinberg, S., Persaud, S. D., Loh, H. H. & Wei, L. N. Stress-induced epigenetic regulation of κ-opioid receptor gene involves transcription factor c-Myc. Proc. Natl Acad. Sci. USA 109, 9167–9172 (2012).
Garcia-Perez, D., Ferenczi, S., Kovacs, K. J. & Milanes, M. V. Distinct regulation pattern of Egr-1, BDNF and Arc during morphine-withdrawal conditioned place aversion paradigm: role of glucocorticoids. Behav. Brain Res 360, 244–254 (2019).
Dietrich, J. B., Takemori, H., Grosch-Dirrig, S., Bertorello, A. & Zwiller, J. Cocaine induces the expression of MEF2C transcription factor in rat striatum through activation of SIK1 and phosphorylation of the histone deacetylase HDAC5. Synapse 66, 61–70 (2012).
Peng, H. Y. et al. Spinal SGK1/GRASP-1/Rab4 is involved in complete Freund’s adjuvant-induced inflammatory pain via regulating dorsal horn GluR1-containing AMPA receptor trafficking in rats. Pain 153, 2380–2392 (2012).
Bawor, M. et al. Contribution of BDNF and DRD2 genetic polymorphisms to continued opioid use in patients receiving methadone treatment for opioid use disorder: an observational study. Addict. Sci. Clin. Pract. 10, 19 (2015).
Sarkar, S., Jain, R., Kethawath, S. M., Gupta, R. & Kumar, M. Serum BDNF levels in patients with opioid dependence during the early withdrawal period: a case control study. Neurosci. Lett. 681, 100–104 (2018).
Gregoretti, I. V., Lee, Y. M. & Goodson, H. V. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338, 17–31 (2004).
Payne, N. C., Maksoud, S., Tannous, B. A. & Mazitschek, R. A direct high-throughput protein quantification strategy facilitates discovery and characterization of a celastrol-derived BRD4 degrader. Cell Chem. Biol. 29, 1333–1340 (2022).
Payne, N. C. & Mazitschek, R. Resolving the deceptive isoform and complex selectivity of HDAC1/2 inhibitors. Cell Chem. Biol. 29, 1140–1152 (2022).
Simonini, M. V. et al. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc. Natl Acad. Sci. USA 103, 1587–1592 (2006).
Stratinaki, M. et al. Regulator of G-protein signaling 4 is a crucial modulator of antidepressant drug action in depression and neuropathic pain models. Proc. Natl Acad. Sci. USA 110, 8254–8259 (2013).
Lutz, P. E. et al. Distinct mu, delta, and kappa opioid receptor mechanisms underlie low sociability and depressive-like behaviors during heroin abstinence. Neuropsychopharmacology 39, 2694–2705 (2014).
Green-Fulgham, S. M. et al. Oxycodone, fentanyl, and morphine amplify established neuropathic pain in male rats. Pain 160, 2634–2640 (2019).
Zhang, Y. et al. Behavioral and neurochemical changes induced by oxycodone differ between adolescent and adult mice. Neuropsychopharmacology 34, 912–922 (2009).
Hoffman, E. M., Watson, J. C., St Sauver, J., Staff, N. P. & Klein, C. J. Association of long-term opioid therapy with functional status, adverse outcomes, and mortality among patients with polyneuropathy. JAMA Neurol. 74, 773–779 (2017).
Comer, S. D., Sullivan, M. A., Vosburg, S. K., Kowalczyk, W. J. & Houser, J. Abuse liability of oxycodone as a function of pain and drug use history. Drug Alcohol Depend. 109, 130–138 (2010).
Darnall, B. D. et al. Patient-centered prescription opioid tapering in community outpatients with chronic pain. JAMA Intern. Med. 178, 707–708 (2018).
Bravo, I. M. et al. Divergent behavioral responses in protracted opioid withdrawal in male and female C57BL/6J mice. Eur. J. Neurosci. 51, 742–754 (2020).
Young, R. & Johnson, D. N. Comparison of routes of administration and time course effects of zacopride and buspirone in mice using an automated light/dark test. Pharm. Biochem Behav. 40, 733–737 (1991).
Fulenwider, H. D. et al. Sex differences in oral oxycodone self-administration and stress-primed reinstatement in rats. Addict. Biol. 25, e12822 (2020).
Collins, D., Reed, B., Zhang, Y. & Kreek, M. J. Sex differences in responsiveness to the prescription opioid oxycodone in mice. Pharm. Biochem. Behav. 148, 99–105 (2016).
Lane, D. A. et al. Region-specific changes in the subcellular distribution of AMPA receptor GluR1 subunit in the rat ventral tegmental area after acute or chronic morphine administration. J. Neurosci. 28, 9670–9681 (2008).
Raehal, K. M. et al. Morphine-induced physiological and behavioral responses in mice lacking G protein-coupled receptor kinase 6. Drug Alcohol Depend. 104, 187–196 (2009).
Matthes, H. W. et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature 383, 819–823 (1996).
Carpenter, M. D., Manners, M. T., Heller, E. A. & Blendy, J. A. Adolescent oxycodone exposure inhibits withdrawal-induced expression of genes associated with the dopamine transmission. Addict. Biol. 26, e12994 (2021).
Sanchez, V., Carpenter, M. D., Yohn, N. L. & Blendy, J. A. Long-lasting effects of adolescent oxycodone exposure on reward-related behavior and gene expression in mice. Psychopharmacol. 233, 3991–4002 (2016).
Markovic, T. et al. Pain induces adaptations in ventral tegmental area dopamine neurons to drive anhedonia-like behavior. Nat. Neurosci. 24, 1601–1613 (2021).
Zhou, H. et al. Inhibition of the prefrontal projection to the nucleus accumbens enhances pain sensitivity and affect. Front. Cell Neurosci. 12, 240 (2018).
Ren, W. et al. The indirect pathway of the nucleus accumbens shell amplifies neuropathic pain. Nat. Neurosci. 19, 220–222 (2016).
Kennedy, P. J. et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat. Neurosci. 16, 434–440 (2013).
Sanna, M. D., Guandalini, L., Romanelli, M. N. & Galeotti, N. The new HDAC1 inhibitor LG325 ameliorates neuropathic pain in a mouse model. Pharm. Biochem Behav. 160, 70–75 (2017).
Borgonetti, V. & Galeotti, N. Combined inhibition of histone deacetylases and BET family proteins as epigenetic therapy for nerve injury-induced neuropathic pain. Pharm. Res. 165, 105431 (2021).
Sanna, M. D. & Galeotti, N. The HDAC1/c-JUN complex is essential in the promotion of nerve injury-induced neuropathic pain through JNK signaling. Eur. J. Pharm. 825, 99–106 (2018).
Graff, J. & Tsai, L. H. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111 (2013).
Quinlan, J., Willson, H. & Grange, K. Hopes and fears before opioid tapering: a quantitative and qualitative study of patients with chronic pain and long-term opioids. Br. J. Pain. 15, 120–128 (2021).
Jantarada, C., Silva, C. & Guimaraes-Pereira, L. Prevalence of problematic use of opioids in patients with chronic noncancer pain: a systematic review with meta-analysis. Pain. Pract. 21, 715–729 (2021).
Smit, T. et al. Anxiety sensitivity and pain intensity independently predict opioid misuse and dependence in chronic pain patients. Psychiatry Res. 294, 113523 (2020).
Throckmorton, D. C., Gottlieb, S. & Woodcock, J. The FDA and the next wave of drug abuse—proactive pharmacovigilance. N. Engl. J. Med 379, 205–207 (2018).
Deacon, R. M. Assessing nest building in mice. Nat. Protoc. 1, 1117–1119 (2006).
Pena, C. J. et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat. Commun. 10, 5098 (2019).
Gaspari, S. et al. Suppression of RGSz1 function optimizes the actions of opioid analgesics by mechanisms that involve the Wnt/β-catenin pathway. Proc. Natl Acad. Sci. USA 115, E2085–E2094 (2018).
Bradner, J. E. et al. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc. Natl Acad. Sci. USA 107, 12617–12622 (2010).
Jochems, J. et al. Antidepressant-like properties of novel HDAC6-selective inhibitors with improved brain bioavailability. Neuropsychopharmacology 39, 389–400 (2014).
Acknowledgements
This study was supported by National Institute of Neurological Disorders and Stroke NS086444 (to V.Z.), R01 NS111351, (to V.Z.), R01NS086444S1 (to R.S.) and National Institute on Drug Abuse P01 DA047233 (to E.J.N. and V.Z.), T32 5T32DA007135-34 (to K.D.P.) and R01NS08644S1 (to K.D.P).
Author information
Authors and Affiliations
Contributions
K.D.P. and R.S. performed experiments, experimental design, data analysis, statistical analysis, manuscript writing and editing; A.R., L.S. and C.P. performed bioinformatic analysis, writing and editing; C.P., V.M., A.T.-B., F.S., A.N., I.G., K.D.P. and R.S. contributed to behavioral experiments; H.K., A.R., R.S., S.V., S.G. and C.J.P. contributed to biochemical experiments and manuscript editing; R.M., J.v.D. and M.J. provided HDAC inhibitors, biochemical analysis and manuscript editing; L.S. and E.N. rigorously revised the manuscript; V.Z. contributed to experimental design and writing and editing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. V.Z. and K.D.P. are inventors on a provisional patent regarding RBC1HI as part of an agreement between the Icahn School of Medicine at Mount Sinai and Regenacy Pharmaceuticals.
Peer review
Peer review information
Nature Neuroscience thanks Theodore Price and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Additional affective behavior assays after oxycodone withdrawal with or without SNI.
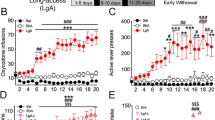
a. No significant difference was observed in the open-field assay (Sham-Sal n = 8, Sham-Oxy n = 9, SNI-Sal n = 8, SNI-Oxy n = 10). b. The time spent in the open arm of the elevated plus maze was significantly increased with oxycodone withdrawal in SNI animals when compared with Sham animals (Sham-Sal n = 9, Sham-Oxy n = 9, SNI-Sal n = 9, SNI-Oxy n = 8; two-way ANOVA interaction F1,31 = 6.299, p = 0.0175; Sidak’s multiple comparisons Oxy SNI vs Sham t = 3.62, df=31, p = 0.0021). c. No significant difference was observed in the nest building assay (Sham-Sal n = 13, Sham-Oxy n = 16, SNI-Sal n = 12, SNI-Oxy n = 14). d. Latency to eat in a novel environment after food deprivation was significantly affected in Sham-Oxy, SNI-Sal, and SNI-Oxy groups as compared with the Sham-Sal controls (Sham-Sal n = 13, Sham-Oxy n = 14, SNI-Sal n = 18, SNI-Oxy n = 15; two-way ANOVA interaction F1,56 = 6.052, p = 0.0170; Sidak’s m.c. SNI-Sal vs Sham-Sal t = 2.963, df=56, p = 0.0089; Sham-Oxy vs Sham-Sal t = 3.249, df=56, p = 0.0039; Tukey’s m.c. SNI-Oxy vs Sham-Sal q = 3.92, df=56, p = 0.0369). e. Total distance traveled during the NSF assay was significantly affected by oxycodone withdrawal in SNI-Oxy and Sham-Oxy (Sham-Sal n = 14, Sham-Oxy n = 13, SNI-Sal n = 12, SNI-Oxy n = 13; two-way ANOVA treatment F1,48 = 20.45, P < 0.001; Sidak’s m.c. Sham-Oxy vs Sham-Sal t = 2.88, df=48, p = 0.0118; SNI-Oxy vs SNI-Sal t = 3.505, df=48, p = 0.002). Values are represented as mean ± SEM *p < 0.05 and ** p < 0.001respectively.
Extended Data Fig. 2 NAc RNA-seq deconvolution across injury and withdrawal conditions.
a–c. RNA-seq deconvolution of Sham-Oxy vs Sham-Saline, SNI-Saline vs Sham Saline, and SNI-Oxy vs Sham-Saline, respectively (n = 4 per group), demonstrating condition-specific alterations in transcriptomic representations of neuronal and vascular cell subtypes (Sham-Oxy vs Sham-Sal Neuron_1 unpaired t-test, t = 2.595, df=6, p = 0.0409; SNI-Sal vs Sham-Sal Mural unpaired t-test, t = 2.456, df=6, p = 0.0494, Neuron_1 unpaired t-test, t = 2.042, df=6, p = 0.0873, Microglia Welch’s unpaired t-test, t = 2.818, df=3.328, p = 0.0593; SNI-Oxy vs Sham-Sal unpaired t-test, t = 2.025, df=6, p = 0.0893). Values are represented as mean ± SEM *p < 0.05.
Extended Data Fig. 3 mPFC and NAc HDAC1 expression in neuronal and glial cells.
a, b. Immunofluorescence staining of HDAC1 in the mPFC and NAc demonstrating high protein nuclear presence across most cells in these regions (naïve mice, n = 3 per group). c, d. RNAscope imaging of Hdac1 transcript colocalization with Tubb3, a pan-neuronal marker, in the mPFC and NAc, demonstrating neuronal expression. e, f. RNAscope imaging of Hdac1 transcript colocalization with Aif1, a microglial marker, demonstrating microglial expression.
Extended Data Fig. 4 Pharmacological and in vivo characterizations of RBC1HI.
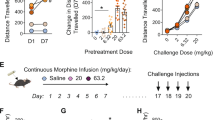
a. Molecular structure of RBC1HI. b. RBC1HI enzymatic inhibition of HDAC1,2,3 (‘HDAC3’=free versus HDAC3 bound to NCOR2) demonstrating highest specificity to HDAC1 (n = 3 replicates; (AUC one-way ANOVA: F3,8 = 191.1, p < 0.0001; Tukey’s m.c. HDAC1 vs HDAC2 q = 11.55, df=8, p = 0.0002; HDAC1 vs free HDAC3 q = 17.22, df=8, p < 0.0001; HDAC1 vs HDAC3/NCOR2 q = 16.21, df=8, p < 0.0001; HDAC2 vs free HDAC3 q = 17.22, df=8, p < 0.0001; HDAC2 vs HDAC3/NCOR2 q = 16.21, df=8, p < 0.0001). c. Plasma bioavailability of RBC1HI at 3 or 10 mg/kg and with intraperitoneal versus oral administration, demonstrating longer bioavailability with the intraperitoneal route. d. Brain bioavailability of RBC1HI with the aforementioned administration doses and routes, demonstrating prolonged drug presence with the intraperitoneal route. Brain to plasma concentration ratios over time. Dose-dependent circulating RBC1HI bioavailability as measured by plasma concentration and brain concentration over time (3 mg/kg i.p.; n = 3 male C57BL/6 mice per time point). e. RBC1HI (3 mg/kg intraperitoneal) did not promote place preference as seen with morphine (6 mg/kg subcutaneous; paired t-test, t = 7.326, d.f. = 4, P = 0.0018). f. 3 mg/kg RBC1HI does not affect locomotor activity up to two hours after administration. Values are represented as mean ± SEM **P < 0.01, ****p < 0.0001.
Extended Data Fig. 5 Additional affective behaviors after treatment with RBC1HI.
a. Habituation phase of Sham-Sal male mice after receiving Veh or RBC1HI, demonstrating equal interaction times for two empty cups (Sham-Sal-Veh n = 9, Sham-Sal-RBC1HI n = 10). b. RBC1HI and Veh animals both showed a preference for social targets in the sociability phase of the social interaction assay (Sham-Sal-Veh n = 8, Sham-Sal-RBC1HI n = 9). c. Female mice undergoing spontaneous withdrawal from oxycodone bury fewer marbles after longitudinal RBC1HI treatment (Sham-Oxy-Veh n = 6, Sham-Oxy-RBC1HI n = 7; unpaired t-test t = 3.663, df=11, p = 0.0037). Values are represented as mean ± SEM **p < 0.01, ****p < 0.0001.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pryce, K.D., Serafini, R.A., Ramakrishnan, A. et al. Oxycodone withdrawal induces HDAC1/HDAC2-dependent transcriptional maladaptations in the reward pathway in a mouse model of peripheral nerve injury. Nat Neurosci 26, 1229–1244 (2023). https://doi.org/10.1038/s41593-023-01350-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-023-01350-3
This article is cited by
-
Tianeptine promotes lasting antiallodynic effects in a mouse model of neuropathic pain
Neuropsychopharmacology (2023)