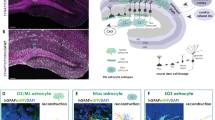
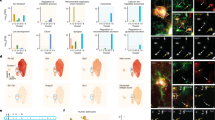
Abstract
Neuronal heterogeneity has been established as a pillar of higher central nervous system function, but glial heterogeneity and its implications for neural circuit function are poorly understood. Here we show that the adult mouse dentate gyrus (DG) of the hippocampus is populated by molecularly distinct astrocyte subtypes that are associated with distinct DG layers. Astrocytes localized to different DG compartments also exhibit subtype-specific morphologies. Physiologically, astrocytes in upper DG layers form large syncytia, while those in lower DG compartments form smaller networks. Astrocyte subtypes differentially express glutamate transporters, which is associated with different amplitudes of glutamate transporter-mediated currents. Key molecular and morphological features of astrocyte diversity in the mice DG are conserved in humans. This adds another layer of complexity to our understanding of brain network composition and function, which will be crucial for further studies on astrocytes in health and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are included in the paper and are available from the corresponding author upon reasonable request. Image source data will be accessible in Figshare (https://figshare.com/s/581158ef2d313e4a027a). The lists of significantly enriched genes in astrocyte subgroups are available as Supplementary Tables 1–5. All data points of all graphs of this study are summarized in Supplementary Table 6.
Code availability
For scRNA-seq/snRNA-seq analysis data were obtained from the GEO repository (GSE95752 (ref. 8) and GSE186538 (ref. 43)). Scripts detailing all aspects of the performed analysis are available as Supplementary Note.
References
Attwell, D. et al. Glial and neuronal control of brain blood flow. Nature 468, 232–243 (2010).
Suzuki, A. et al. Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 144, 810–823 (2011).
Chung, W. S., Allen, N. J. & Eroglu, C. Astrocytes control synapse formation, function, and elimination. Cold Spring Harb. Perspect. Biol. 7, a020370 (2015).
Takata, N. & Hirase, H. Cortical layer 1 and layer 2/3 astrocytes exhibit distinct calcium dynamics in vivo. PLoS ONE 3, e2525 (2008).
Olsen, M. L. et al. New Insights on astrocyte ion channels: critical for homeostasis and neuron-glia signaling. J. Neurosci. 35, 13827–13835 (2015).
Isokawa, M. & McKhann, G. M. II Electrophysiological and morphological characterization of dentate astrocytes in the hippocampus. J. Neurobiol. 65, 125–134 (2005).
Matthias, K. et al. Segregated expression of AMPA-type glutamate receptors and glutamate transporters defines distinct astrocyte populations in the mouse hippocampus. J. Neurosci. 23, 1750–1758 (2003).
Hochgerner, H., Zeisel, A., Lonnerberg, P. & Linnarsson, S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 21, 290–299 (2018).
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014 (2018).
Boisvert, M. M., Erikson, G. A., Shokhirev, M. N. & Allen, N. J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285 (2018).
Buosi, A. S., Matias, I., Araujo, A. P. B., Batista, C. & Gomes, F. C. A. Heterogeneity in synaptogenic profile of astrocytes from different brain regions. Mol. Neurobiol. 55, 751–762 (2018).
Chai, H. et al. Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron 95, 531–549 (2017).
Batiuk, M. Y. et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220 (2020).
Bayraktar, O. A. et al. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509 (2020).
Blanco-Suarez, E., Liu, T. F., Kopelevich, A. & Allen, N. J. Astrocyte-secreted chordin-like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron 100, 1116–1132 (2018).
Molofsky, A. V. et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194 (2014).
Farmer, W. T. et al. Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 351, 849–854 (2016).
Kempermann, G., Song, H. & Gage, F. H. Neurogenesis in the adult hippocampus. Cold Spring Harb. Perspect. Med. 5, a018812 (2015).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Beyer, F., Lüdje, W., Karpf, J., Saher, G. & Beckervordersandforth, R. Distribution of Aldh1L1-CreER(T2) recombination in astrocytes versus neural stem cells in the neurogenic niches of the adult mouse brain. Front. Neurosci. 15, 713077 (2021).
Schneider, J. et al. Astrogenesis in the murine dentate gyrus is a life-long and dynamic process. EMBO J. 41, e110409 (2022).
Ohlig, S. et al. Molecular diversity of diencephalic astrocytes reveals adult astrogenesis regulated by Smad4. EMBO J. 40, e107532 (2021).
Bohrer, C. et al. The balance of Id3 and E47 determines neural stem/precursor cell differentiation into astrocytes. EMBO J. 34, 2804–2819 (2015).
Nolte, C. et al. GFAP promoter-controlled EGFP-expressing transgenic mice: a tool to visualize astrocytes and astrogliosis in living brain tissue. Glia 33, 72–86 (2001).
Kosaka, T. & Hama, K. Three-dimensional structure of astrocytes in the rat dentate gyrus. J. Comp. Neurol. 249, 242–260 (1986).
Bushong, E. A., Martone, M. E. & Ellisman, M. H. Examination of the relationship between astrocyte morphology and laminar boundaries in the molecular layer of adult dentate gyrus. J. Comp. Neurol. 462, 241–251 (2003).
Ogata, K. & Kosaka, T. Structural and quantitative analysis of astrocytes in the mouse hippocampus. Neuroscience 113, 221–233 (2002).
Beckervordersandforth, R. et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7, 744–758 (2010).
Hirrlinger, J. et al. Split-Cre complementation indicates coincident activity of different genes in vivo. PLoS ONE 4, e4286 (2009).
Dallérac, G., Zapata, J. & Rouach, N. Versatile control of synaptic circuits by astrocytes: where, when and how? Nat. Rev. Neurosci. 19, 729–743 (2018).
Anders, S. et al. Spatial properties of astrocyte gap junction coupling in the rat hippocampus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130600 (2014).
Danbolt, N. C. Glutamate uptake. Prog. Neurobiol. 65, 1–105 (2001).
Rose, C. R. et al. Astroglial glutamate signaling and uptake in the hippocampus. Front Mol. Neurosci. 10, 451 (2017).
Perego, C. et al. The GLT-1 and GLAST glutamate transporters are expressed on morphologically distinct astrocytes and regulated by neuronal activity in primary hippocampal cocultures. J. Neurochem. 75, 1076–1084 (2000).
Batiuk, M. Y. et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J. Biol. Chem. 292, 8874–8891 (2017).
Kantzer, C. G. et al. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia 65, 990–1004 (2017).
Benraiss, A. et al. Cell-intrinsic glial pathology is conserved across human and murine models of Huntington’s disease. Cell Rep. 36, 109308 (2021).
Li, J. et al. Conservation and divergence of vulnerability and responses to stressors between human and mouse astrocytes. Nat. Commun. 12, 3958 (2021).
Eckenhoff, M. F. & Rakic, P. Radial organization of the hippocampal dentate gyrus: a Golgi, ultrastructural, and immunocytochemical analysis in the developing rhesus monkey. J. Comp. Neurol. 223, 1–21 (1984).
Franjic, D. et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron 110, 452–469 (2022).
Szabo, Z. et al. Extensive astrocyte synchronization advances neuronal coupling in slow wave activity in vivo. Sci. Rep. 7, 6018 (2017).
Schlett, K. Glutamate as a modulator of embryonic and adult neurogenesis. Curr. Top. Med. Chem. 6, 949–960 (2006).
Guo, Y. et al. The effects of astrocytes on differentiation of neural stem cells are influenced by knock-down of the glutamate transporter, GLT-1. Neurochem. Int. 63, 498–506 (2013).
Oberheim, N. A. et al. Uniquely hominid features of adult human astrocytes. J. Neurosci. 29, 3276–3287 (2009).
Han, X. et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell 12, 342–353 (2013).
Nakamura, T., Colbert, M. C. & Robbins, J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 98, 1547–1554 (2006).
Beckervordersandforth, R. et al. In vivo targeting of adult neural stem cells in the dentate gyrus by a split-cre approach. Stem Cell Rep. 2, 153–162 (2014).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Breithausen, B., Kautzmann, S., Boehlen, A., Steinhauser, C. & Henneberger, C. Limited contribution of astroglial gap junction coupling to buffering of extracellular K+ in CA1 stratum radiatum. Glia 68, 918–931 (2020).
Fischer, J. et al. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat. Protoc. 6, 1981–1989 (2011).
Dietrich, J. & Kempermann, G. Role of endogenous neural stem cells in neurological disease and brain repair. Adv. Exp. Med Biol. 557, 191–220 (2006).
Moreno-Jiménez, E. P. et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlage, 2016).
Acknowledgements
We thank C. Lie, M. Llorens-Martin and V. Piatti for fruitful discussions. We thank J. Winkler as a member of the JK’s TAC committee for constructive comments. This work was supported by grants from the German Research Foundation (DFG; SPP1757 BE5136/2-1, BE5136/1-2 to R.B., INST 410/45-1 FUGG; SFB1089 B03, SPP1757 HE6949/1, FOR2795 and HE6949/3 to C.H., Emmy Noether Program grant 455354162 to A.S.), by the Johannes-and Frieda-Marohn Stiftung to F.B., by the LOEWE CePTER Epilepsy Research Center of the state Hessen to E.F. and S.L. and by the Excellence Cluster Cardio-Pulmonary Institute to S.L., Bavarian State Ministry of Sciences, Research, and the Arts (ForInter; F.2-F2412.30/1/24) and IZKF (P074) to S.F. The DFG research training group 2162 “Neurodevelopment and Vulnerability of the Central Nervous System” supported this work as follows: J.K., M.T.-W. and N.C. as fellows and J.S. as an associated fellow. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The present work was performed in the fulfillment of the requirements for obtaining the degree, “Dr. med”.
Author information
Authors and Affiliations
Contributions
J.K., C.H. and R.B. were responsible for conceptualization; J.K., P.U., N.C., F.B., M.-T. W., J.S., E.F., S.F., A.S. and R.B. were responsible for investigation; J.K., P.U., N.C., F.B., M.-T. W., J.S., E.F., S.F., A.S. and R.B. were responsible for formal analysis; A.R., J.B., S.B., S.L., S.F., A.S., C.H. and R.B. were responsible for resources and funding acquisition; R.B. was responsible for writing the original draft; A.R., S.L., C.H. and R.B were responsible for supervision. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors declare that the submitted work was performed without any personal, professional or financial relationships that could be potentially construed as a conflict of interest.
Peer review
Peer review information
Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 scRNA-seq revealed molecularly diverse astrocyte subtypes associated to distinct DG layers.
(a) Heatmap showing differentially regulated genes in the five clusters. Differential gene analysis was performed by using the Wilcoxon Rank sum test as implemented in Seurat. (b) Examples of genes expressed by all DG astrocytes as represented by tSNE plots and immunostaining against the transcription factors ID4 and SOX2 (in white) and GFAP (magenta). (c) tSNE plots of DEGs upregulated in cluster 4 (circle) revealed many known markers for radial glia-like NSCs. Immunostaining against NESTIN (white) and GFAP (magenta) confirmed the expression in radial glia-like NSCs. (d) scRNA seq analysis revealed that S100ß, Aldh1L1 and Aqp4 are expressed in all DG astrocytes except radial glia-like NSCs (circle), which was confirmed by immunostaining against the markers (white) and GFAP (magenta). For AQP4, hGFAPeGFP animals were used to delineate whole astrocytes based on the cytoplasmic GFP signal. In the left panel, the arrows indicate expression of AQP4 in a GZ astrocyte surrounding a blood vessel. The right panel depicts a GFP+ radial glial-like NSCs that lack AQP4 expression at its end feet (arrows). (e) tSNE plots depicting the expression of genes associated to gliogenesis (Id3), cell cycle (Ccnd2, Pcna), and BMP signaling (Bmp1, Jun) in cluster 2 (circle) indicating that cluster 2 represents proliferating astrocytes. (f) Confocal images of dividing astrocytes in the adult mouse DG expressing cell cycle marker MCM2 (white) and astrocyte markers GFAP (magenta) and SOX2 (blue) in hGFAPeGFP animals. (g) tSNE blots of DEG upregulated in cluster 3 (circle) in comparison to cluster 0 and 1. Cluster 3 represented SGZ astrocytes as verified by spatial expression (Fig. 1i-k; Fig. 4b, d) and functional analysis of Glutamate transporter currents (Fig. 4e–g). (h) tSNEs of DEGs upregulated in cluster 0 in comparison to cluster 1, including Slc38a3 and Fam107a, for which RNAscope analysis was performed (Fig. 1l, n). All immunostainings were performed in 4–7 experimental animals in at least 3 independent experiments; all scale bars = 20 µm.
Extended Data Fig. 2 Morphologically distinct astrocyte subtypes localized to different layers of the adult DG.
(a) Confocal images of a hGFAPeGFP brain slice stained only with DAPI (blue) representing an overview of astrocytes subtypes localized to different DG layers; more than 10 experimental animals were analyzed. (b) Heatmap of 3D-reconstructed astrocytes from all DG layers; subtypes are depicted on the right side and marked by color code (as depicted in Fig. 2q, r); morphological parameters depicted below revealed clustering of astrocytes populating the same DG layer. (c-l) Graphs representing the individual morphological parameters used to conduct the PCA. Astrocyte subtypes are indicated below the graphs; red dots represent analysed cells; 8 experimental animals were analysed for morphological analysis. Data are represented as mean ± SEM; one-way ANOVA with Tukey post hoc tests were performed to determine significance; no adjustments were made for multiple comparison; in case of significance, p-values are indicated in the graphs. Scale bars = 20 µm.
Extended Data Fig. 3 Astrocyte subtypes were embedded in a layer-specific cellular environment and displayed subtype-specific gap junction-coupling.
(a) Quantification of astrocytes associated to different DG layers. (b, c) Immunostaining and quantification of IBA+ microglia (red) per DG compartment; DAPI (blue) (d, e) Immunostaining and quantification of PDGFRα+ (red) per DG compartment. 3–6 experimental animals were analyzed and are indicated as red dots representing biological replicates. (f) Schematic representation of Alexa Fluor 594 dye-injections into astrocytes in acute hippocampal slices. (g-j) Two-photon excitation images of astrocyte subtypes from hGFAPeGFP mice. Brightfield images in the upper row show localisation of dye-injected astrocyte subtypes within the adult DG; lower row depicts Alexa Fluor 594 diffusion to gap junction-coupled cells; astrocyte subtypes as indicated. (j) Cellular layers were identified using infrared image channel using DIC (left panel), and astrocytes were classified according to their location to an identified layer. Dye-coupled astrocytes were identified using two-photon excitation imaging of AlexaFluor 594 diffusing from the patched cell (right panel; green line represents SGZ). (k) Representative analysis of a single experiment. A set of dye-coupled cells (dots) and a monoexponential fit (I(d) = 100 % * exp(−d/Cλ), where I is the somatic fluorescence intensity, d is the three-dimensional distance to the patched cell, and Cλ the coupling length constant34. (I) Residuals of this representative example (k) of a monoexponential fit of dye-coupled cells. (m) Residual values of all monoexponential fits for dye-loaded hilus astrocytes (n = 7 patched astrocytes); total of 61 coupled cells (58 hilus astrocytes in red and 3 SGZ astrocytes in blue). Boxed area: same residuals to visualize their spread around zero. (n) Residual values of monoexponential fits for dye-loaded SGZ astrocytes (n = 8 patched astrocytes); total of 46 coupled cell (17 hilus astrocytes in red and 29 SGZ astrocytes in blue). Boxed area: same residuals to visualize their spread around zero. (o) Residual values of monoexponential fits for dialysed ML astrocytes (n = 5 patched astrocytes); total of 50 coupled cells (40 ML astrocytes in green and 10 SGZ/hilar astrocyte in red). Boxed area: same residuals to visualize their spread around zero. Data are represented as mean ± SEM. One-way ANOVA plus Tukey post-hoc tests were performed to determine statistical significance; no adjustments were made for multiple comparison; p-values are indicated in the graphs. Scale bares = 20 µm (b, d), and 50 µm (g-j).
Extended Data Fig. 4 Isolation of distinct astrocytes subtypes by FACS and astrocyte diversity in the adult human DG.
(a) Dot blots depicting FACS gating strategy of adult DG cells according to living (left) and single cells (middle); isotype-matched controls conjugated to PE (a) and APC (b) were used to set the gates for sorting of ACSA1 (SLC1A3)-APC and ACSA2 (ATP1B2)-PE (right). (c) Graph depicting diameter of primary neurospheres generated by SLC1A3+/ATP1B2+ cells, ATP1B2+ cells, SLC1A31+ cells, and the all negative-sorted cell fractions; no significant differences were detected between the population. Neurosphere assay was carried out from 4 independent FACS experiments (3–6 wildtype mice per experiment). One-way ANOVA with Tukey post hoc tests were performed to determine significance; no adjustments were made for multiple comparisons. (d-i) High magnification images of hippocampal sections from three human subjects between 43 and 52 years of age (represent patient 1–3 in average intensity blots Fig. 4h, j, l) were stained against GFAP (magenta), EAATs (white) and DAPI (blue). Immunostaining against SLC1A3 (d-f) and SLC1A2 (g-i). Immunostainings against each marker were performed in 3 patients in 3 independent experiments. Scale bars = 20 µm.
Extended Data Fig. 5 Common and divergent transcriptomic signatures of mouse and human DG astrocyte subtypes.
(a) Top 30 DEGs for each human DG astrocyte cluster. (b) Heatmap depicting pseudobulk gene expression levels for the same genes as in (a). Color bar for each row depicts if the gene was identified as differentially expressed in the pseudobulk differential gene expression analysis. (c) Percentage of DEGs detected by Seurat that were also detected in the pseudobulk differential gene expression analysis either for all detected genes (red bars) or the top 30 DEGs (cyan bars). Differential gene analysis was performed by using the Wilcoxon Rank sum test as implemented in Seurat. Pseudobulk differential gene expression analysis was performed using glmQLFTest function implemented in edgeR.
Extended Data Fig. 6 Common and divergent transcriptomic signatures of mouse and human DG astrocyte subtypes.
(a–f) Examples of genes differentially expressed in mouse niche astrocytes (a, b), radial glia-like NSCs (c) and proliferating astrocytes (d–f) as represented by tSNE plots (upper row) and UMAP plots representing their expression pattern in human DG astrocytes (lower row). (g-j) ISH images from the Allen Mouse Brain Atlas (g-i; mouse.brain-map.org) and Allen Human Brain Atlas (j; human.brain-map.org) for Glul (mouse.brain-map.org/ish/experiment/show/14421), Grm3 (mouse.brain-map.org/ish/experiment/show/72231), Slc1a3 (mouse.brain-map.org/ish/experiment/show/20274), SLC1A3 (human.brain-map.org/ish/experiment/show/159113739). (k-p) Analysis of the integrated mouse and human data sets revealed candidates showing a corresponding expression in mouse (dark blue, upper row) and human (red, lower row) astrocytes belonging to the same clusters. (q) CTNNA2 was predominantly expressed only in human astrocytes and (r) LDHB in mouse astrocytes. (s-u) Immunostaining against HOPX (white), GFAP (magenta), and DAPI in human sections. (s, t) HOPX is not expressed in adult hippocampal and cortical astrocytes in postmortem slices (s; arrow) and in freshly fixed adult DG tissue (t). (u) However, almost all cortical astrocytes expressed HOPX in the fetal brain (arrowheads). HOPX expression was determined in 3 independent experiments in 5 human individuals. All scale bares = 20 µm, except (g-i) = 50 µm, and (j) = 100 µm; UMAP = Uniform Manifold Approximation and Projection; gw = gestational week.
Supplementary information
Supplementary Information
Supplementary Note—Workflow of human to mice scRNA-seq/snRNA-seq data comparison.
Supplementary Table 1
Differentially expressed genes of clusters 0–4 compared to all others.
Supplementary Table 2
Differentially expressed genes upregulated in cluster 3 in comparison to clusters 0 and 1.
Supplementary Table 3
Differentially expressed genes of clusters 0 and 1 compared to each other.
Supplementary Table 4
Differentially expressed genes of human DG astrocyte clusters identified by Seurat.
Supplementary Table 5
Differentially expressed genes of human DG astrocyte clusters identified by pseudobulk differential gene expression analysis using EdgeR.
Supplementary Table 6
Summary table containing statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karpf, J., Unichenko, P., Chalmers, N. et al. Dentate gyrus astrocytes exhibit layer-specific molecular, morphological and physiological features. Nat Neurosci 25, 1626–1638 (2022). https://doi.org/10.1038/s41593-022-01192-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01192-5
This article is cited by
-
Astrocytes in the adult dentate gyrus—balance between adult and developmental tasks
Molecular Psychiatry (2024)
-
Host brain environmental influences on transplanted medial ganglionic eminence progenitors
Scientific Reports (2024)