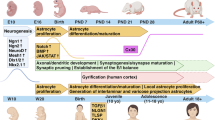
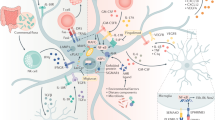
Abstract
Astrocytes negatively impact neuronal development in many models of neurodevelopmental disorders (NDs); however, how they do this, and if mechanisms are shared across disorders, is not known. In this study, we developed a cell culture system to ask how astrocyte protein secretion and gene expression change in three mouse models of genetic NDs (Rett, Fragile X and Down syndromes). ND astrocytes increase release of Igfbp2, a secreted inhibitor of insulin-like growth factor (IGF). IGF rescues neuronal deficits in many NDs, and we found that blocking Igfbp2 partially rescues inhibitory effects of Rett syndrome astrocytes, suggesting that increased astrocyte Igfbp2 contributes to decreased IGF signaling in NDs. We identified that increased BMP signaling is upstream of protein secretion changes, including Igfbp2, and blocking BMP signaling in Fragile X and Rett syndrome astrocytes reverses inhibitory effects on neurite outgrowth. This work provides a resource of astrocyte-secreted proteins in health and ND models and identifies novel targets for intervention in diverse NDs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA sequencing raw data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE139285. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride) with the dataset identifier PXD015996. Source data that support all findings in the study are available as supplementary tables (proteomics and RNA sequencing) and source data tables (statistical analysis). The data that support this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
No new code was used in this study.
References
Ebrahimi-Fakhari, D. & Sahin, M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Curr. Opin. Neurol. 28, 91–102 (2015).
Amir, R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999).
Xu, X., Miller, E. C. & Pozzo-Miller, L. Dendritic spine dysgenesis in Rett syndrome. Front. Neuroanat. 8, 97 (2014).
Wang, I. T. J., Reyes, A.-R. S. & Zhou, Z. Neuronal morphology in MeCP2 mouse models is intrinsically variable and depends on age, cell type, and Mecp2 mutation. Neurobiol. Dis. 58, 3–12 (2013).
Irwin, S. A., Galvez, R. & Greenough, W. T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex 10, 1038–1044 (2000).
Nimchinsky, E. A., Oberlander, A. M. & Svoboda, K. Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci. 21, 5139–5146 (2001).
Benavides-Piccione, R. et al. On dendrites in Down syndrome and DS murine models: a spiny way to learn. Prog. Neurobiol. 74, 111–126 (2004).
Blanco-Suárez, E., Caldwell, A. L. M. & Allen, N. J. Role of astrocyte–synapse interactions in CNS disorders. J. Physiol. 595, 1903–1916 (2017).
Banker, G. Trophic interactions between astroglial cells and hippocampal neurons in culture. Science 209, 809–810 (1980).
Ullian, E. M., Sapperstein, S. K., Christopherson, K. S. & Barres, B. A. Control of synapse number by glia. Science 291, 657–661 (2001).
Araujo, B. H. S., et al. Down syndrome iPSC-derived astrocytes impair neuronal synaptogenesis and the mTOR pathway in vitro. Mol. Neurobiol. 55, 5962–5975 (2017).
Ballas, N., Lioy, D. T., Grunseich, C. & Mandel, G. Non–cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat. Neurosci. 12, 311–317 (2009).
Jacobs, S., Nathwani, M. & Doering, L. Fragile X astrocytes induce developmental delays in dendrite maturation and synaptic protein expression. BMC Neurosci. 11, 132 (2010).
Meissner, F., Scheltema, R. A., Mollenkopf, H.-J. & Mann, M. Direct proteomic quantification of the secretome of activated immune cells. Science 340, 475–478 (2013).
Johnson, E. C. B. et al. Large-scale deep multi-layer analysis of Alzheimer’s disease brain reveals strong proteomic disease-related changes not observed at the RNA level. Nat. Neurosci. 25, 213–225 (2022).
Boisvert, M. M., Erikson, G. A., Shokhirev, M. N. & Allen, N. J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285 (2018).
Chai, H. et al. Neural circuit-specialized astrocytes: transcriptomic, proteomic, morphological, and functional evidence. Neuron 95, 531–549 (2017).
McCarthy, K. D. & de Vellis, J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85, 890–902 (1980).
Foo, L. C. et al. Development of a method for the purification and culture of rodent astrocytes. Neuron 71, 799–811 (2011).
Foo, L. C. Purification of rat and mouse astrocytes by immunopanning. Cold Spring Harb. Protoc. 2013, 421–432 (2013).
Batiuk, M. Y. et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J. Biol. Chem. 292, 8874–8891 (2017).
Kantzer, C. et al. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia 65, 990–1004 (2017).
Steinmetz, C. C., Buard, I., Claudepierre, T., Nägler, K. & Pfrieger, F. W. Regional variations in the glial influence on synapse development in the mouse CNS. J. Physiol. 577, 249–261 (2006).
Williams, E. C. et al. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum. Mol. Genet. 23, 2968–2980 (2014).
Allen, N. J. & Eroglu, C. Cell biology of astrocyte–synapse interactions. Neuron 96, 697–708 (2017).
Yang, Y. H., Nam, M. S. & Yang, E. S. Rapid prenatal diagnosis of trisomy 21 by real-time quantitative polymerase chain reaction with amplification of small tandem repeats and S100B in chromosome 21. Yonsei Med. J. 46, 193–197 (2005).
Higashimori, H. et al. Selective deletion of astroglial FMRP dysregulates glutamate transporter GLT1 and contributes to fragile X syndrome phenotypes in vivo. J. Neurosci. 36, 7079–7094 (2016).
Higashimori, H. et al. Astroglial FMRP-dependent translational down-regulation of mGluR5 underlies glutamate transporter GLT1 dysregulation in the fragile X mouse. Hum. Mol. Genet. 22, 2041–2054 (2013).
Yuskaitis, C. J., Beurel, E. & Jope, R. S. Evidence of reactive astrocytes but not peripheral immune system activation in a mouse model of fragile X syndrome. Biochim. Biophys. Acta 1802, 1006–1012 (2010).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481 (2017).
Blanco-Suarez, E., Liu, T.-F., Kopelevich, A. & Allen, N. J. Astrocyte-secreted chordin-like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron 100, 1116–1132.e1113 (2018).
Molofsky, A. V. et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194 (2014).
Scholze, A. R., Foo, L. C., Mulinyawe, S. & Barres, B. A. BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS ONE 9, e110668 (2014).
Yang, Q. et al. Excessive astrocyte-derived neurotrophin-3 contributes to the abnormal neuronal dendritic development in a mouse model of fragile X syndrome. PLoS Genet. 8, e1003172 (2012).
Vilardell, M. et al. Meta-analysis of heterogeneous Down syndrome data reveals consistent genome-wide dosage effects related to neurological processes. BMC Genomics 12, 229 (2011).
Ballestín, R. et al. Astrocytes of the murine model for Down syndrome Ts65Dn display reduced intracellular ionic zinc. Neurochem. Int. 75, 48–53 (2014).
Costales, J. & Kolevzon, A. The therapeutic potential of insulin-like growth factor-1 in central nervous system disorders. Neurosci. Biobehav. Rev. 63, 207–222 (2016).
Woronowicz, A. et al. Carboxypeptidase E knockout mice exhibit abnormal dendritic arborization and spine morphology in central nervous system neurons. J. Neurosci. Res. 88, 64–72 (2010).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Chen, C. et al. Role of astroglia in Down’s syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat. Commun. 5, 4430 (2014).
Marchetto, M. C. N. et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539 (2010).
Wang, R. N. et al. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105 (2014).
Verkhratsky, A., Matteoli, M., Parpura, V., Mothet, J.-P. & Zorec, R. Astrocytes as secretory cells of the central nervous system: idiosyncrasies of vesicular secretion. EMBO J. 35, 239–257 (2016).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Bayraktar, O. A. et al. Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509 (2020).
Kashima, R. et al. Augmented noncanonical BMP type II receptor signaling mediates the synaptic abnormality of fragile X syndrome. Sci. Signal. 9, ra58 (2016).
Chen, J., Alberts, I. & Li, X. Dysregulation of the IGF-I/PI3K/AKT/mTOR signaling pathway in autism spectrum disorders. Int. J. Dev. Neurosci. 35, 35–41 (2014).
Izumi, K. et al. Elevation of insulin-like growth factor binding protein-2 level in Pallister–Killian syndrome: Implications for the postnatal growth retardation phenotype. Am. J. Med. Genet. A 167, 1268–1274 (2015).
Itoh, M. et al. Methyl CpG-binding protein 2 (a mutation of which causes Rett syndrome) directly regulates insulin-like growth factor binding protein 3 in mouse and human brains. J. Neuropathol. Exp. Neurol. 66, 117–123 (2007).
Degano, A. L., Pasterkamp, R. J. & Ronnett, G. V. MeCP2 deficiency disrupts axonal guidance, fasciculation, and targeting by altering semaphorin 3F function. Mol. Cell. Neurosci. 42, 243–254 (2009).
Menon, L. & Mihailescu, M.-R. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 35, 5379–5392 (2007).
Abouzied, M. M., El-Tahir, H. M., Gieselmann, V. & Franken, S. Hepatoma-derived growth factor-related protein-3: a new neurotrophic and neurite outgrowth-promoting factor for cortical neurons. J. Neurosci. Res. 88, 3610–3620 (2010).
Kalus, I. et al. Sulf1 and Sulf2 differentially modulate heparan sulfate proteoglycan sulfation during postnatal cerebellum development: evidence for neuroprotective and neurite outgrowth promoting functions. PLoS ONE 10, e0139853 (2015).
Bao, X. et al. Heparin-binding growth factor, pleiotrophin, mediates neuritogenic activity of embryonic pig brain-derived chondroitin sulfate/dermatan sulfate hybrid chains. J. Biol. Chem. 280, 9180–9191 (2005).
Krencik, R. et al. Dysregulation of astrocyte extracellular signaling in Costello syndrome. Sci. Transl. Med. 7, 286ra266 (2015).
Malter, J. S., Ray, B. C., Westmark, P. R. & Westmark, C. J. Fragile X sndrome and Alzheimer’s disease: another story about APP and beta-amyloid. Curr. Alzheimer Res 7, 200–206 (2010).
Seritan, A. L. et al. Dementia in fragile X-associated tremor/ataxia syndrome (FXTAS): comparison with Alzheimer’s disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 1138–1144 (2008).
Bonham, L. W. et al. Insulin-like growth factor binding protein 2 is associated with biomarkers of alzheimer’s disease pathology and shows differential expression in transgenic mice. Front. Neurosci. 12, 476 (2018).
Guy, J., Hendrich, B., Holmes, M., Martin, J. E. & Bird, A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27, 322–326 (2001).
Bakker, C. E. et al. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell 78, P23–P33 (1994).
Gupta, M., Dhanasekaran, A. R. & Gardiner, K. J. Mouse models of Down syndrome: gene content and consequences. Mamm. Genome 27, 538–555 (2016).
Davisson, M. T., Schmidt, C. & Akeson, E. C. Segmental trisomy of murine chromosome 16: a new model system for studying Down syndrome. Prog. Clin. Biol. Res. 360, 263–280 (1990).
Duchon, A. et al. Identification of the translocation breakpoints in the Ts65Dn and Ts1Cje mouse lines: relevance for modeling Down syndrome. Mamm. Genome 22, 674–684 (2011).
Kahlem, P. et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 14, 1258–1267 (2004).
Yu, T. et al. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum. Mol. Genet. 19, 2780–2791 (2010).
Bansal, R. & Pfeiffer, S. E. Reversible inhibition of oligodendrocyte progenitor differentiation by a monoclonal antibody against surface galactolipids. Proc. Natl Acad. Sci. USA 86, 6181–6185 (1989).
Farhy-Tselnicker, I. et al. Astrocyte-secreted glypican 4 regulates release of neuronal pentraxin 1 from axons to induce functional synapse formation. Neuron 96, 428–445.e413 (2017).
Winzeler, A. & Wang, J. T. Purification and culture of retinal ganglion cells from rodents. Cold Spring Harb. Protoc. 2013, 643–652 (2013).
Xu, T. et al. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteom. 129, 16–24 (2015).
Carvalho, P. C. et al. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat. Protoc. 11, 102–117 (2016).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 (2006).
Mi, H. et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 (2017).
Farhy-Tselnicker, I. et al. Activity-dependent modulation of synapse-regulating genes in astrocytes. eLife 10, e70514 (2021).
Acknowledgements
We thank C. Dowling for technical assistance and A. Saghatelian, members of the Allen laboratory and Molecular Neurobiology Laboratory (MNL) for helpful discussions. This work was supported by a Salk Institute Innovation grant. A.L.M.C. was supported by a Dennis Weatherstone Predoctoral Fellowship from Autism Speaks and the Chapman Foundation. J.D. is supported by F30 HD106699 from the National Institute of Child Health and Human Development. N.J.A. is supported by the Chan Zuckerberg Initiative, the Hearst Foundation and the Pew Foundation. This work was supported by core facilities of the Salk Institute, including the Mass Spectrometry Core, the Next Generation Sequencing Core, the Integrative Genomics and Bioinformatics Core and the Waitt Advanced Biophotonics Core, with funding from National Institutes of Health–National Cancer Institute Cancer Center Support Grant (P30 014195), the Chapman Foundation, the Waitt Foundation and the Helmsley Center for Genomic Medicine. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
A.L.M.C. and N.J.A. designed the experiments and wrote the manuscript, with input from all other authors. N.J.A. conceived the project. A.L.M.C., N.J.A., J.D., L.S., A.B., A.M. and J.K.D. performed experiments. N.J.A., A.L.M.C., J.K.D., J.D. and M.N.S. analyzed data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Giovanna Mallucci and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Immunopanned astrocyte and neuron cultures for study of NDs.
a,b. Immunostaining IP-astrocyte cultures for cell type markers reveals that the majority of cells express astrocyte-associated proteins Gfap and Aqp4, while rarely expressing NeuN (neuronal marker), Iba1 (microglial marker) or NG2 (oligodendrocyte precursor cell marker), N = 15 experiments (3WT, 1 RTT, 3 FXS, 8 DS; no differences were observed between ND and WT expression of cell markers). c. Example images of WT immunopanned neurons cultured for 2 days in WT ACM, and immunostained for MAP2 to identify neurons and antibodies against cortical neuron layer-enriched marker genes. Data analyzed in Fig. 1g. N = 3 experiments SATB2, TBR1, CTIP2; N = 4 Reelin; N = 2 GAD. d. Example images from Fig. 1h, prior to processing and analysis. WT neurons immunostained with MAP2 (dendrites, green) and tau (axon, red). e. Relative frequency of total neurite outgrowth, example experiment shown, same data as Fig. 1i. f. Examination of additional measures of neurite growth for experiments in Fig. 1h,i. Violin plot: dashed line median, dotted lines 25th and 75th percentile. Number of neurons: alone = 51, WT ACM = 75. Statistics by 2-sided Mann-Whitney test.
Extended Data Fig. 2 IP astrocytes reproduce in vivo alterations to ND astrocytes.
a. qRT-PCR for cell-type markers from mRNA collected from IP astrocytes compared to P7 mouse cortex demonstrates enrichment for astrocytes (Gfap), a depletion of neurons (Syt1), microglia (Cd68), fibroblasts (Fgfr4), and a decrease in oligodendrocyte precursor cells (OPCs; Cspg4) in WT and ND IP astrocyte cultures. N = 6 cultures per genotype. WT data same as in Fig. 1b. b. Reactive astrocyte markers from pan reactive, inflammation-induced and injury-induced reactive astrocytes are not consistently altered in IP astrocyte cultures from ND compared to WT, demonstrating cultures are not reactive. Data from RNA sequencing. N = 6 cultures WT, RTT, FXS; 4 DS. * adjusted p < 0.05, FPKM > 1 and fold change ≥1.5 calculated with DESeq2.
Extended Data Fig. 3 Altered protein secretion and gene levels of ND astrocytes.
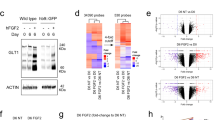
a,c,e. Volcano plot of genes expressed by astrocytes at FPKM > 1, plotted as Log2FC against -log10 p-value comparing each disorder to WT: FXS (a), RTT (c) and DS (e). Each dot represents a gene; top right sector genes significantly upregulated (FC > 1.5, adjusted p-value <0.05), examples in red; top left sector genes significantly downregulated (FC < -1.5, adjusted p-value <0.05), examples in blue, calculated with DESeq2. b,d,f. Venn diagram of overlap between proteins and genes with decreased level in FXS (b), RTT (d) and DS (f) ACM and astrocytes g,h. Venn diagram showing overlap in proteins downregulated in all ND (g), and heatmap of top altered proteins ranked by abundance in WT ACM (h). i,j. Venn diagram showing number of genes downregulated (i) and corresponding heatmap (j) of overlapping altered genes. Scale bar in j also applies to heatmap in h. k. Pathway analysis of proteins and genes altered in ND astrocytes compared to WT demonstrates some overlapping and some unique alterations in ND astrocyte function compared to WT, performed using PANTHER over-representation test with Fisher’s Exact test and FDR corrected. Proteomics, N = 6 cultures per genotype, p < 0.05, abundance >0.01%, fold change between WT and ND ≥ 1.5. RNASeq, N = 6 cultures WT, RTT, FXS; 4 DS, adjusted p < 0.05, FPKM > 1, fold change between ND and WT ≥ 1.5.
Extended Data Fig. 4 Excess Igfbp2 in ACM inhibits neurite outgrowth.
a. Expression of IGF family members in cortical cell types (data from Zhang et al., 2014). b,c. Addition of Igfbp2 protein to WT ACM inhibits WT neurite outgrowth, which is reduced by adding IGF1. Addition of CPE protein to WT ACM does not inhibit WT neurite outgrowth. b. Example images of WT neurons cultured for 48 hours, conditions as marked (image merge of MAP2 + Tau). c. Quantification of total neurite outgrowth. Example experiment shown, repeated 2 times with same result, number of neurons: control alone=49, control ACM = 50, CPE alone=36, CPE ACM = 46, Igfbp2 alone=44, Igfbp2 ACM = 48, Igfbp2 ACM + Igf1 = 39. d. smFISH against Igfbp2 mRNA in the P7 visual cortex in Aldh1l1-GFP mice to mark astrocytes, combined with probe for OPCs (Cspg4). See Fig. 4g for quantification. N = 3 WT mice. e. Example images from Fig. 4d prior to processing and analysis. Neurons immunostained with MAP2 (dendrites, green) and tau (axon, red). f. Relative frequency distribution plot of total neurite outgrowth length, pooled data from 3 experiments, same data as Fig. 4e. g. Adding the IgG control antibody to WT ACM does not alter neurite outgrowth. Example experiment shown, repeated twice with same result. Number of neurons: control alone=216, control ACM = 333, Igfbp2-Ab alone=257, Igfbp2-Ab ACM = 267, IgG con-Ab alone=277, IgG con-Ab ACM = 266. Violin plots (c,g), dashed line marks median, dotted lines 25th and 75th percentile. Statistics by Kruskal-Wallis one-way ANOVA on ranks with Dunn’s test for multiple comparisons, p values compared to control alone condition (c).
Extended Data Fig. 5 Blocking Igfbp2 in RTT reduces neural developmental deficits.
a,c,e. Example images from Fig. 5a,c,e prior to processing and analysis. Neurons cultured for 48 hours in RTT (a), FXS (c) or DS (e) ACM and immunostained with MAP2 (dendrites, green) and tau (axon, red). b,d,f. Relative frequency distribution plot of total neurite outgrowth, pooled data from 3 (b), 4 (d), 5 (f) experiments per graph, same data as Fig. 5b, d, f. Number of neurons: RTT (b): alone=439, WT ACM = 549, RTT ACM = 633, RTT ACM + Igfbp2-Ab=621; FXS (d): alone=492, WT ACM = 608, FXS ACM = 621, FXS ACM + Igfbp2-Ab=647; DS (f): alone=716, WT ACM = 992, DS ACM = 765, DS ACM + Igfbp2-Ab=858. g. Relative frequency distribution plot of neuronal cell body size, pooled data from 3 experiments, same data as Fig. 5g. h-l Cell body size of upper layer cortical neurons is not different between WT and RTT mice, and unaffected by the Igfbp2 neutralizing antibody. h. Schematic of the experiment: P2 mice were injected in the visual cortex with AAV synapsin-GFP + /- antibody, and tissue collected at P7 and GFP-expressing neurons imaged, with the region to be imaged identified by the presence of the fluorescently labeled antibody. i,j. Cell body area of upper layer neurons is unaltered in RTT compared to WT mice. i. Analysis by mice, graph average ± s.e.m., individual data points represent mice, N = 4 WT and 4 RTT mice, statistics by 2-sided T-test. j. Analysis by cells, graph individual data points represent cells, dashed line represents the mean, n = 162 WT and 177 RTT cells, statistics by 2-sided T-test. k,l. Cell body area of upper layer neurons in RTT mice is unaffected by an Igfbp2 neutralizing antibody. k. Analysis by mice, graph average ± s.e.m., individual data points represent mice, N = 5 control-Ab and 5 Igfbp2-Ab mice, statistics by 2-sided T-test. l. Analysis by cells, graph individual data points represent cells, dashed line represents the mean, n = 183 control-Ab and 195 Igfbp2-Ab cells, statistics by 2-sided T-test.
Extended Data Fig. 6 Activating WT astrocyte BMP signaling mimics ND astrocytes.
a. Relative expression of BMP family members in purified cell types in the cortex shows enrichment for BMP target genes in astrocytes (data from Zhang et al, 2014). b,c. BMP6-treated astrocytes have thinner more branched processes (b), and increased expression of both GFAP (cyan) and AQP4 (magenta) (c). Example images shown, experiment repeated 3 times with same effect. d. Volcano plot of genes expressed by astrocytes at FPKM > 1, plotted as Log2FC against -log10 p-value comparing BMP6-treated and untreated WT astrocytes. Each dot represents a gene; top right sector genes significantly upregulated (FC > 1.5, adjusted p-value <0.05), examples in red; top left sector genes significantly downregulated (FC < -1.5, adjusted p-value <0.05), examples in blue, calculated with DESeq2. e. Venn diagram showing overlap between proteins and genes downregulated in WT astrocytes treated with BMP6. For mass spectrometry and RNA Sequencing N = 6 cultures, half of each culture treated with BMP6 and other half left untreated. Proteomics, p < 0.05, abundance >0.01%, fold change ≥1.5 calculated with Patternlab. RNASeq, adjusted p < 0.05, FPKM > 1, fold change ≥1.5 calculated with DESeq2.
Extended Data Fig. 7 Blocking ND astrocyte BMP signaling reduces neural deficits.
a,b. BMP6-treated WT astrocytes show protein secretion (a) and gene expression (b) downregulations that overlap with ND astrocytes. N = 6 cultures WT, FXS, RTT, DS, plus 6 cultures WT + /- BMP6 proteomics; N = 6 cultures WT, FXS, RTT; 4 DS, plus 6 cultures WT + /- BMP6 RNA sequencing. c. Example images from Fig. 7d prior to processing and analysis. Neurons immunostained with MAP2 (dendrites, green) and tau (axon, red). d. Relative frequency distribution plot of total neurite outgrowth length, same data as Fig. 7e. Data from 3 experiments, number of neurons: alone=467, WT ACM = 733, BMP6 WT ACM = 610.e. Relative frequency distribution plot of total neurite outgrowth length, same data as Fig. 7 f. Data from 3 experiments, number of neurons: alone=378, WT ACM = 380, BMP6 WT ACM = 506, BMP6 WT ACM + Igfbp2 blocking Ab=335. f. Example images from Fig. 7h prior to processing and analysis. Neurons immunostained with MAP2 (dendrites, green) and tau (axon, red). g. Relative frequency distribution plot of total neurite outgrowth, same data as Fig. 7i. Data from 3 experiments, number of neurons: alone=923, WT ACM = 1164, FXS ACM = 1132, Noggin FXS ACM = 1099. h. Relative frequency distribution plot of total neurite outgrowth, same data as Fig. 7j. Data from 3 experiments, number of neurons: alone=238, WT ACM = 279, RTT ACM = 387, Noggin RTT ACM = 365. i. Example images of cortical neurons treated with noggin at the time of plating, ± WT ACM or ± FXS ACM (image merge of MAP2 + Tau). j. Quantification of total neurite outgrowth, data from 3 experiments. Number of neurons: alone=2197, alone + noggin=1981, WT ACM = 2167, WT ACM + noggin=2421, FXS ACM = 2060, FXS ACM + noggin=2523. Violin plots dashed line marks median, dotted lines 25th and 75th percentile. Statistics by Kruskal-Wallis one-way ANOVA on ranks with Dunn’s test for multiple comparisons.
Supplementary information
Supplementary Information
Supplementary Methods
Supplementary Table 1
Gene expression profiles and statistical comparisons of acutely isolated IP astrocytes (IP A), cultured IP astrocytes (IP C) and cultured MD isolated astrocytes (MD C) presented as TPM. Statistics were calculated with DESeq2.
Supplementary Table 2
Proteomic profiles of ACM from WT, RTT, FXS, DS and BMP6-treated astrocytes in vitro. Spectral count data were normalized as NSAF and presented as % abundance; a cutoff was set at 0.01% of total protein to be included.
Supplementary Table 3
Gene expression profiles and statistical comparisons of WT, RTT, FXS, DS and BMP6-treated astrocytes in vitro, presented as FPKM. Statistics were calculated with DESeq2.
Supplementary Table 4
Genes triplicated in TS65Dn mouse and expression level in DS astrocytes; overlap with upregulated genes and proteins in DS astrocytes. Proteomics statistics were calculated with PatternLab; RNA sequencing statistics were calculated with DESeq2.
Supplementary Table 5
Change in secreted proteins between FXS, RTT, DS and BMP6-treated ACM compared to WT, as well as overlapping changes among all three NDs and all three NDs and BMP6-treated ACM. Protein annotation from UniProt for proteins altered in all three NDs compared to WT. Statistics were calculated with PatternLab.
Supplementary Table 6
Pathway analysis of secreted protein changes between FXS, RTT, DS and BMP6-treated ACM compared to WT. Calculated with PANTHER over-representation test with Fisher’s exact test and FDR corrected.
Supplementary Table 7
Changes in gene expression between FXS, RTT, DS, and BMP6-treated astrocytes compared to WT as well as all overlapping changes between all three NDs and BMP6-treated astrocytes compared to WT. Statistics were calculated with DESeq2.
Supplementary Table 8
Pathway analysis of gene expression changes in FXS, RTT, DS and BMP6-treated astrocytes compared to WT, calculated with PANTHER over-representation test with Fisher’s exact test and FDR corrected
Supplementary Table 9
Overlapping protein secretion and gene expression changes between FXS, RTT, DS and BMP6-treated astrocytes compared to WT. Proteomics statistics were calculated with PatternLab; RNA sequencing statistics were calculated with DESeq2.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Fig. 6
Statistical source data
Source Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 7
Statistical source data
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Caldwell, A.L.M., Sancho, L., Deng, J. et al. Aberrant astrocyte protein secretion contributes to altered neuronal development in multiple models of neurodevelopmental disorders. Nat Neurosci 25, 1163–1178 (2022). https://doi.org/10.1038/s41593-022-01150-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01150-1