Abstract
Hydrocephalus, characterized by cerebral ventricular dilatation, is routinely attributed to primary defects in cerebrospinal fluid (CSF) homeostasis. This fosters CSF shunting as the leading reason for brain surgery in children despite considerable disease heterogeneity. In this study, by integrating human brain transcriptomics with whole-exome sequencing of 483 patients with congenital hydrocephalus (CH), we found convergence of CH risk genes in embryonic neuroepithelial stem cells. Of all CH risk genes, TRIM71/lin-41 harbors the most de novo mutations and is most specifically expressed in neuroepithelial cells. Mice harboring neuroepithelial cell-specific Trim71 deletion or CH-specific Trim71 mutation exhibit prenatal hydrocephalus. CH mutations disrupt TRIM71 binding to its RNA targets, causing premature neuroepithelial cell differentiation and reduced neurogenesis. Cortical hypoplasia leads to a hypercompliant cortex and secondary ventricular enlargement without primary defects in CSF circulation. These data highlight the importance of precisely regulated neuroepithelial cell fate for normal brain–CSF biomechanics and support a clinically relevant neuroprogenitor-based paradigm of CH.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Resource availability
All unique animal lines and cell lines in this study are available upon reasonable request to the corresponding author. Source data are provided with this paper.
Data availability
Unprocessed scans of all immunoblots in the paper are included as Source Data Fig. 1. WES data have been deposited in the database of Genotypes and Phenotypes under accession number phs000744. A Life Sciences Reporting Summary is available for this paper. In-house pipelines are available from the corresponding author upon reasonable request. Bulk RNA sequencing data of mESCs have been deposited at the Gene Expression Omnibus under accession number GSE189420. Source data are provided with this paper.
Code availability
All code used in this paper is available from the corresponding author upon reasonable request.
References
Aristotle. The History of Animals http://classics.mit.edu/Aristotle/history_anim.mb.txt
Duy, P. Q. et al. Brain ventricles as windows into brain development and disease. Neuron 110, 12–15 (2022).
Persson, E. K., Hagberg, G. & Uvebrant, P. Disabilities in children with hydrocephalus—a population-based study of children aged between four and twelve years. Neuropediatrics 37, 330–336 (2006).
Tully, H. M. & Dobyns, W. B. Infantile hydrocephalus: a review of epidemiology, classification and causes. Eur. J. Med. Genet. 57, 359–368 (2014).
Greenberg, M. S. Handbook of Neurosurgery, 9th edition (Thieme, 2019).
Furey, C. G. et al. De novo mutation in genes regulating neural stem cell fate in human congenital hydrocephalus. Neuron 99, 302–314 (2018).
Jin, S. C. et al. Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus. Nat. Med. 26, 1754–1765 (2020).
Kousi, M. & Katsanis, N. The genetic basis of hydrocephalus. Annu. Rev. Neurosci. 39, 409–435 (2016).
Huang, J. K. et al. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. 6, 484–495 (2018).
Willsey, H. R. et al. Parallel in vivo analysis of large-effect autism genes implicates cortical neurogenesis and estrogen in risk and resilience. Neuron 109, 788–804 (2021).
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Li, M\. et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615 (2018).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Nowakowski, T. J. et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017).
Willsey, A. J. et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013).
Silbereis, J. C., Pochareddy, S., Zhu, Y., Li, M. & Sestan, N. The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268 (2016).
Mizrak, D. et al. Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 26, 394–406 (2019).
DeSisto, J. et al. Single-cell transcriptomic analyses of the developing meninges reveal meningeal fibroblast diversity and function. Dev. Cell 54, 43–59 (2020).
Parikshak, N. N. et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Duy, P. Q., Furey, C. G. & Kahle, K. T. Trim71/lin-41 links an ancient miRNA pathway to human congenital hydrocephalus. Trends Mol. Med. 25, 467–469 (2019).
Spassky, N. et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 25, 10–18 (2005).
Coletti, A. M. et al. Characterization of the ventricular-subventricular stem cell niche during human brain development. Development 145, dev170100 (2018).
Maller Schulman, B. R. et al. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7, 3935–3942 (2008).
Chen, J., Lai, F. & Niswander, L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 26, 803–815 (2012).
Mitschka, S. et al. Co-existence of intact stemness and priming of neural differentiation programs in mES cells lacking Trim71. Sci. Rep. 5, 11126 (2015).
Nguyen, D. T. T. et al. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ. 24, 1063–1078 (2017).
Dong, J. et al. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 19, 31 (2018).
Burke, E. E. et al. Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. Nat. Commun. 11, 462 (2020).
Onorati, M. et al. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 16, 2576–2592 (2016).
Dubois, N. C., Hofmann, D., Kaloulis, K., Bishop, J. M. & Trumpp, A. Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis 44, 355–360 (2006).
Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002).
Worringer, K. A. et al. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14, 40–52 (2014).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Steffensen, A. B. et al. Cotransporter-mediated water transport underlying cerebrospinal fluid formation. Nat. Commun. 9, 2167 (2018).
Karimy, J. K. et al. Inflammation-dependent cerebrospinal fluid hypersecretion by the choroid plexus epithelium in posthemorrhagic hydrocephalus. Nat. Med. 23, 997–1003 (2017).
Date, P. et al. Visualizing flow in an intact CSF network using optical coherence tomography: implications for human congenital hydrocephalus. Sci. Rep. 9, 6196 (2019).
Roossien, D. H., Miller, K. E. & Gallo, G. Ciliobrevins as tools for studying dynein motor function. Front. Cell Neurosci. 9, 252 (2015).
Peña, A., Harris, N. G., Bolton, M. D., Czosnyka, M. & Pickard, J. D. Communicating hydrocephalus: the biomechanics of progressive ventricular enlargement revisited. Acta Neurochir. Suppl. 81, 59–63 (2002).
Duy, P. Q. & Kahle, K. T. Intraventricular cerebrospinal fluid turbulence in pediatric communicating hydrocephalus. Neurology 97, 246–247 (2021).
Elkin, B. S., Ilankovan, A. & Morrison, B. Age-dependent regional mechanical properties of the rat hippocampus and cortex. J. Biomech. Eng. 132, 011010 (2010).
Canovic, E. P. et al. Characterizing multiscale mechanical properties of brain tissue using atomic force microscopy, impact indentation, and rheometry. J. Vis. Exp. 54201 (2016).
Yalcin, S. E. et al. Electric field stimulates production of highly conductive microbial OmcZ nanowires. Nat. Chem. Biol. 16, 1136–1142 (2020).
Torres-Fernández, L. A. et al. The mRNA repressor TRIM71 cooperates with nonsense-mediated decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Res. 47, 11861–11879 (2019).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Chang, H.-M. M. et al. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 3, 923 (2012).
Welte, T. et al. The RNA hairpin binder TRIM71 modulates alternative splicing by repressing MBNL1. Genes Dev. 33, 1221–1235 (2019).
Phoenix, T. N. & Temple, S. Spred1, a negative regulator of Ras-MAPK-ERK, is enriched in CNS germinal zones, dampens NSC proliferation, and maintains ventricular zone structure. Genes Dev. 24, 45–56 (2010).
DeSpenza, T. et al. PTEN mutations in autism spectrum disorder and congenital hydrocephalus: developmental pleiotropy and therapeutic targets. Trends Neurosci. 44, 961–976 (2021).
van der Linden, V. et al. Association of severe hydrocephalus with congenital Zika syndrome. JAMA Neurol. 76, 203 (2019).
Su, J. et al. Novel compound heterozygous frameshift variants in WDR81 associated with congenital hydrocephalus 3 with brain anomalies: first Chinese prenatal case confirms WDR81 involvement. Mol. Genet. Genom. Med. 9, e1624 (2021).
Cavallin, M. et al. WDR81 mutations cause extreme microcephaly and impair mitotic progression in human fibroblasts and Drosophila neural stem cells. Brain 140, 2597–2609 (2017).
Sullivan, W. et al. Exome sequencing as a potential diagnostic adjunct in sporadic congenital hydrocephalus. JAMA Pediatr. 175, 310 (2021).
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735 (2009).
Duran, D. et al. Mutations in chromatin modifier and ephrin signaling genes in vein of Galen malformation. Neuron 101, 429–443 (2019).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 11.10.1–11.10.33 (2013).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907 (2012).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Gudmundsson, S. et al. Addendum: the mutational constraint spectrum quantified from variation in 141,456 humans. Nature 597, E3–E4 (2021).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Samocha, K. E. et al. Regional missense constraint improves variant deleteriousness prediction. Preprint at https://www.biorxiv.org/content/10.1101/148353v1 (2017).
Dong, C. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137 (2015).
Wei, Q. et al. A Bayesian framework for de novo mutation calling in parents–offspring trios. Bioinformatics 31, 1375–1381 (2015).
Jin, S. C. et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49, 1593–1601 (2017).
Robinson, J. T., Thorvaldsdóttir, H., Wenger, A. M., Zehir, A. & Mesirov, J. P. Variant review with the Integrative Genomics Viewer. Cancer Res. 77, e31–e34 (2017).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Ware, J. S., Samocha, K. E., Homsy, J. & Daly, M. J. Interpreting de novo variation in human disease using denovolyzeR. Curr. Protoc. Hum. Genet. 87, 7.25.1–7.25.15 (2015).
Jin, S. C. et al. Mutations disrupting neuritogenesis genes confer risk for cerebral palsy. Nat. Genet. 52, 1046–1056 (2020).
Piñero, J. et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839 (2017).
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 (2005).
Walker, R. L. et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 179, 750–771 (2019).
Yang, H., Wang, H. & Jaenisch, R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 9, 1956–1968 (2014).
Chen, S., Lee, B., Lee, A. Y.-F., Modzelewski, A. J. & He, L. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem. 291, 14457–14467 (2016).
Behringer, R., Gertsenstein, M., Nagy, K. V. & Nagy, A. Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2014).
Nichols, J. & Jones, K. Derivation of mouse embryonic stem (ES) cell lines using small-molecule inhibitors of Erk and Gsk3 signaling (2i). Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot094086 (2017).
Sbalzarini, I. F. & Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151, 182–195 (2005).
Chenouard, N. et al. Objective comparison of particle tracking methods. Nat. Methods 11, 281–289 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Benhalevy, D., McFarland, H. L., Sarshad, A. A. & Hafner, M. PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites. Methods 118–119, 41–49 (2017).
Acknowledgements
This work was supported by T32GM136651 (to P.Q.D.), F30HD106694 (to P.Q.D.), RO1NS111029 (to K.T.K.), David M. Rubenstein Fund for Hearing Research (to A. Doetzlhofer), 1R21NS121642-01 (to X.Y.), 1R01NS122904-01 (to X.Y.), the Rudi Schulte Institute (to K.T.K.), the Hydrocephalus Association Innovator Award (to K.T.K., E.D. and S.J.C.), 5R21NS116484-02 (to E.D.), National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI) Pathway to Independence award R00HL143036-02 (to S.J.C.), Clinical & Translational Research Funding Program award CTSA1405 (to S.J.C.), Children’s Discovery Institute Faculty Scholar award CDI-FR-2021-926 (to S.J.C.), NIH Director’s New Innovator award 1DP2AI138259-01 (to N.S.M.), the Career Award at the Scientific Interfaces from Burroughs Welcome Fund (to N.S.M.), the Hartwell Foundation Individual Biomedical Research Award (to N.S.M.), MacBrainResource NIH MH113257 (to A. Duque), NIH DA023999 (to P.R.) and Deutsche Forschungsgemeinschaft (German Research Foundation) award WU 563/3-1 (to F.G.W.). W.K. is funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy EXC2151–390873048. All mechanical measurements were performed at the Yale West Campus Imaging Core. CSF outflow imaging was performed using the small animal imaging facility of the Center for Precision Cancer Modeling, sponsored by the Yale Cancer Center. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Certain cartoons and diagrams were generated using BioRender.
Author information
Authors and Affiliations
Contributions
Conceptualization of the project: P.Q.D., S.C.W., W.K. and K.T.K. Data collection: P.Q.D., S.C.W., C.M., X.L., D.L., P.J.D., S.M., A.S., W.D., J.J., E.K., A.J.K., S.K., M.Z.P., L.A.T.-F., K.H., E.D.D., M.B., T.U., S.A.J., E.C., L.T.H., B.J., A.M.M.S., F.L., S.-K.K., M.L., Y.Y., Y.T., A. Duque, C.N.-W., Y.H., K.S., S.M.R., A.K.S., G.A., C.G.F., A.T.T., B.C.R., H.S., A. Dunbar, T.D., J.G., A.M., A.M.-D.-L., X.Y., W.E.B., B.S.C., E.M.R.L., R.T.C., P.R., H.L., E.D., H.B., N.M., J.I.E.-V., C.A.W., S.L.A., J.S., K.P., A. Doetzlhofer, F.G.W., S.C.J., R.P.L., N.S., W.K. and K.T.K. Data analysis: P.Q.D., S.C.W., C.M., X.L., D.L., P.J.D., S.M., A.S., W.D., J.J., E.K., A.J.K., S.K., M.Z.P., L.A.T.-F., K.H., E.D.D., M.B., T.U., S.A.J., E.C., L.T.H., B.J., A.M.M.S., F.L., S.-K.K., M.L., Y.Y., Y.T., A. Duque, C.N.-W., Y.H., K.S., S.M.R., A.K.S., G.A., C.G.F., A.T.T., B.C.R., H.S., A. Dunbar, T.D., J.G., A.M., A.M.-D.-L., X.Y., W.E.B., B.S.C., E.M.R.L., R.T.C., P.R., H.L., E.D., H.B., N.M., J.I.E.-V., C.A.W., S.L.A., J.S., K.P., A. Doetzlhofer, F.G.W., S.C.J., R.P.L., N.S., W.K. and K.T.K. Writing: P.Q.D., S.C.W., W.K. and K.T.K., with input from all authors.
Corresponding author
Ethics declarations
Competing interests
J.J. is an employee of GeneDx, Inc.
Peer review
Peer review information
Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Integrative genomic analysis of CH genetic risk.
a) Heatmap showing expression of CH risk genes across developmental timepoints and regions of the developing human brain. Analyzed transcriptomic dataset from12. b) Enrichment of CH risk genes across developmental timepoints of the human brain in a microarray dataset of the human brain from11. PCW: post-conception week, M: month, Y: year. Boxplot (in f): median (line), 25th and 75th percentiles (box), whiskers extend up to 1.5 times the interquartile range from the top (bottom) of the box to the furthest datum within that distance. Significance was calculated by comparing to background expression using one-sided Wilcoxon rank sum test. For detailed statistical information, see Supplementary Table 13. c-e) CH risk gene network connectivity in different layers of the prenatal human frontal neocortex. Analyzed transcriptomic dataset from13. c) VZ- and SVZi-specific CH risk gene interaction networks have the highest average connectivity per interaction. d-e) VZ (d) and SVZi (e) have significantly higher CH risk gene interaction network connectivity than expected by chance. The red line indicates the observed connectivity, and the gray histogram shows the null distribution from 1,000 permutations. P values were calculated by permutation tests (see Methods). f) Heatmap showing maximal expression of CH risk genes across different cell types of the developing human cortex. Analyzed transcriptomic dataset from12. g) Enrichment of CH risk genes in a prenatal human brain single-cell RNA sequencing data set from14. Purple square highlights lack of enrichment in choroid plexus cells. Choroid: choroid plexus, Glyc: glycolysis, IPC: intermediate progenitor cells, MGE: medial ganglionic eminence, OPC: oligodendrocyte precursor cells, RGC: radial glia cells. P values were calculated by hypergeometric test. h) Enrichment of CH risk genes in a postnatal mouse ventricular wall single-cell RNA sequencing data set from17. Purple square highlights lack of enrichment in ependymal cells. aNSC: actively dividing neural stem cells, TAC: transit amplifying cells, NB: neuroblasts, OPC: oligodendrocyte progenitor cells, COP: committed oligodendrocyte precursors. P values were calculated by hypergeometric test. i) Enrichment of CH risk genes in a prenatal mouse meninges single-cell RNA sequencing data set from18. P values were calculated by hypergeometric tests.
Extended Data Fig. 2 Gene co-expression network analysis of CH genetic risk.
a) Clustering dendrogram of genes showing module membership in colors. The y axis represents network distance as determined by 1-topological overlap (TO), where values closer to 0 indicate greater similarity of probe expression. Analyzed transcriptomic dataset from12. b-g) GO term enrichment analysis of gene co-expression modules that are enriched for CH risk genes. Significance was calculated by two-sided Fisher’s exact test. h-i) Co-expression network connectivity between CH and other developmental disorder risk genes. CH risk genes exhibit greater connectivity to cortical developmental disorder genes than to PCD genes. Significance was tested using one-sided Wilcoxon rank sum test by comparing to the reference group, CH-PCD.
Extended Data Fig. 3
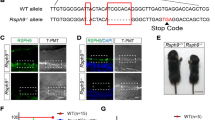
Macrocephalic appearance of a hydrocephalic Trim71R595H/+ mouse compared to a WT control at P21.
Extended Data Fig. 4 Characterization of TRIM71 expression.
a) TRIM71 is minimally expressed in the adult human cortex. Analyzed single-nucleus RNA-sequencing data from12. b-c) RNAscope in situ hybridization of Trim71 in mouse embryonal carcinoma cells (b) or mouse Foxj1-expressing ependymal cell cultures (c) d) TRIM71 immunostaining in the WT mouse choroid plexus at P7.
Extended Data Fig. 5 Conditional knockout of Trim71 in embryonic NSCs by Nestin-Cre results in hydrocephalus in a subset of mice.
a) A subset of Nestin-Trim71fl/fl mice develop hydrocephalus with obvious macrocephaly at P21. b) Brain MRI demonstrates severe ventriculomegaly at P21 in a representative hydrocephalic Nestin-Cre;Trim71fl/fl mouse compared to a Trim71fl/fl control. 3D reconstructions of the ventricular system based on MRI scans are shown.
Extended Data Fig. 6 Mutant TRIM71 leads to reduced cell proliferation upon neural differentiation and an altered transcriptome.
a) Schematic of experimental paradigm of eFluor670 proliferation assay. Fluorescence intensity decreases by half upon every cell division. b,c) Representative histograms of eFluor670-labelled mESCs undergoing neural differentiation. Proliferation defects in mutant (Trim71R595H/+, Trim71R595H/R595H) (b) and Trim71 KO (c) mESC are reflected by slower loss of fluorescence intensity over time than that in WT or Trim71fl/fl control mESCs. d) Heatmap of the bulk RNA-sequencing showing gene expression profiles clustered in modules over mutant Trim71 genotypes.
Extended Data Fig. 7 Low levels of active caspase 3 (aCASP3) staining in the brains of control and Trim71 mutant mouse models.
a) aCASP3 staining in the neural tube neuroepithelia of WT, Trim71R595H/+, and Trim71R595H/R595H embryos at E9.5. b) aCASP3 staining in the cortices and lateral ventricles of WT and hydrocephalic Trim71R595H/+ mice at P0. c) aCASP3 staining in the cortices and lateral ventricles of Trim71fl/fl and hydrocephalic Nestin-Cre;Trim71fl/fl mice at P0.
Extended Data Fig. 8
H&E stained images of histological sections throughout the rostrocaudal extent of the midbrain cerebral aqueduct in P0 hydrocephalic Nestin-Trim71fl/fl mouse and a Trim71fl/fl control, demonstrating anatomical patency of the aqueduct in both genotypes.
Extended Data Fig. 9 Characterization of ependymal cilia and choroid plexus in hydrocephalic Trim71 mutant mice.
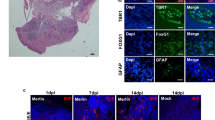
a) Coronal brain sections from control and hydrocephalic mice were stained for ciliary markers (acetyl-α-tubulin and ARL13B), ependymal cell markers (S100B and FOXJ1), and choroid plexus epithelial cell markers (E-Cadherin, OTX2).b-d) Immunostaining for molecular correlates of choroid plexus hypersecretion in WT controls and hydrocephalic Trim71R595H/+ P0 mice. Immunostaining of phosphorylated SLC12A2/NKCC1 (pSLC12A2) (b) and phosphorylated STK39/SPAK (pSTK39) (c) in the choroid plexi of a WT control and hydrocephalic Trim71R595H/+ P0 mouse. d) Immunostaining of SLC12A2/NKCC1 and STK39/SPAK in the choroid plexi of a WT control and hydrocephalic Trim71R595H/+ P0 mouse. e-f) Validation of OCT imaging platform to characterize ependymal cilia-driven CSF flow ex vivo. e). Representative flow polarity maps demonstrating bead trajectories (by temporal color coding) over time in an adult WT mouse brain explant in toxin-naive condition, with ciliobrevin, and after washout of toxin. Color bar represents color versus corresponding frame in the color-coded image. f) Quantitation of local CSF flow speed at the ventricular wall in brain explants from adult WT mouse brains in the different experimental conditions. g-i) CSF flow directionality in WT littermate controls and hydrocephalic Trim71R595H/+ mice at P7. g) Post-Gaussian processing CSF flow maps in a WT littermate control and hydrocephalic Trim71R595H/+ mice at P7. h,i) Quantitation of CSF flow directionality in WT littermate controls and hydrocephalic Trim71R595H/+ mice at P7. Directionality is represented as the distance between the start point and end point (d) divided by the bead pathway (d), see panel (i). Significance was tested by a two-sided paired t-test (f) or two-sided unpaired t-test (h): *: P<0.05, **: P<0.01, ns (not significant): P>0.05. Data represented as boxplots (f,h): median (line), 25th and 75th percentiles (box), whiskers go down to the smallest value and up to the largest, overlaid with individual data points. For detailed statistical information (f, h), see Supplementary Table 13.
Extended Data Fig. 10 TRIM71 CH-mutations lead to deregulation of Cdkn1a/p21 and Egr1 and loss of interaction to the NMD factor UPF1.
a,b) Representative immunoblots for the CLIP-qPCR assays in a) HEK293T cells and b) mESC. c-e) Protein levels of EGR1 and P21/CDKN1A in WT and mutant TRIM71 mESC. c) Representative immunoblots showing protein levels of EGR1 and P21/CDKN1A. d,e) Quantitation of EGR1 (d) and P21/CDKN1A (e) protein levels from immunoblots in (c). f-i) De-repression of CDKN1A 3′UTR in HEK293T cells overexpressing TRIM71 mutants and in Trim71 mutant mESCs. f) Schematic of luciferase assay to examine RNA target silencing by TRIM71 (CDKN1A as example). g) Luciferase reporter assay for the CDKN1A 3′UTR showing repression ability of the indicated TRIM71 constructs in HEK293T cells. Norm. RLU = normalized relative light units. h) Representative immunoblot showing TRIM71 construct overexpression for the CDKN1A-3′UTR luciferase assays in (g). i) Luciferase reporter assay for the CDKN1A 3′UTR showing repression ability of the indicated mESC line. j,k) CH-causing mutations impair TRIM71 binding to the NMD factor UPF1. Immunoblots show UPF1 enrichment upon co-precipitation with j) different Ig-tagged TRIM71 constructs transfected into HEK293T cells, namely Ig-Ctrl, Ig-TRIM71, Ig-ΔNHL6, Ig-R608H, and Ig-R796H or k) with endogenous FLAG-tagged TRIM71 and TRIM71-R595H in mESC. Statistical significance was tested by a two-sided, one-sample t-test (d, e, g, i): *: P<0.05, **: P<0.01, ***: P<0.001. Data represented as mean±s.e.m., overlaid with individual data points. For detailed statistical information (d, e, g, i), see Supplementary Table 13. Source data are provided.
Supplementary information
Supplementary Video 1
OCT imaging of cilia-generated CSF flow in a mouse adult ventricular explant in naive condition (top), with ciliobrevin toxin (middle) and after ciliobrevin toxin washout (bottom). The same ventricular explant was imaged for all three conditions.
Source data
Source Data Fig. 7
Unprocessed immunoblots in Fig. 7.
Source Data Extended Data Fig. 10
Unprocessed immunoblots in Extended Data Fig. 10.
Rights and permissions
About this article
Cite this article
Duy, P.Q., Weise, S.C., Marini, C. et al. Impaired neurogenesis alters brain biomechanics in a neuroprogenitor-based genetic subtype of congenital hydrocephalus. Nat Neurosci 25, 458–473 (2022). https://doi.org/10.1038/s41593-022-01043-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-022-01043-3
This article is cited by
-
The homozygous pathogenic variant of the POMGNT1 gene identified using whole-exome sequencing in Iranian family with congenital hydrocephalus
Egyptian Journal of Medical Human Genetics (2024)
-
The genetic basis of hydrocephalus: genes, pathways, mechanisms, and global impact
Fluids and Barriers of the CNS (2024)
-
A year in review: brain barriers and brain fluids research in 2022
Fluids and Barriers of the CNS (2023)
-
Heterozygous FOXJ1 Mutations Cause Incomplete Ependymal Cell Differentiation and Communicating Hydrocephalus
Cellular and Molecular Neurobiology (2023)
-
Gold nanoparticle-enhanced X-ray microtomography of the rodent reveals region-specific cerebrospinal fluid circulation in the brain
Nature Communications (2023)