Abstract
Behaviors and disorders related to self-regulation, such as substance use, antisocial behavior and attention-deficit/hyperactivity disorder, are collectively referred to as externalizing and have shared genetic liability. We applied a multivariate approach that leverages genetic correlations among externalizing traits for genome-wide association analyses. By pooling data from ~1.5 million people, our approach is statistically more powerful than single-trait analyses and identifies more than 500 genetic loci. The loci were enriched for genes expressed in the brain and related to nervous system development. A polygenic score constructed from our results predicts a range of behavioral and medical outcomes that were not part of genome-wide analyses, including traits that until now lacked well-performing polygenic scores, such as opioid use disorder, suicide, HIV infections, criminal convictions and unemployment. Our findings are consistent with the idea that persistent difficulties in self-regulation can be conceptualized as a neurodevelopmental trait with complex and far-reaching social and health correlates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data sources are described in the Supplementary Information and are listed in the Reporting Summary. No new data were collected. Only data from existing studies or study cohorts were analyzed, some of which have restricted access to protect the privacy of the study participants (see Reporting Summary for accession codes or URLs). The minimum dataset necessary to interpret, verify and extend the research, that is, the GWAS summary statistics for the EXT GWAS (our main discovery analysis), can be obtained by following the procedures detailed at https://externalizing.org/request-data/. In brief, summary statistics are derived from analyses based in part on 23andMe data, for which we are restricted to only publicly available report results for up to 10,000 SNPs. The full set of externalizing GWAS summary statistics can be made available to qualified investigators who enter into an agreement with 23andMe that protects participant confidentiality. Once the request has been approved by 23andMe, a representative of the Externalizing Consortium can share the full GWAS summary statistics.
References
Richmond-Rakerd, L. S. et al. Clustering of health, crime and social-welfare inequality in 4 million citizens from two nations. Nat. Hum. Behav. 4, 255–264 (2020).
Case, A. & Deaton, A. Mortality and morbidity in the 21st century. Brookings Pap. Econ. Act. 2017, 397–476 (2017).
Achenbach, T. M. The classification of children’s psychiatric symptoms: a factor-analytic study. Psychol. Monogr. 80, 1–37 (1966).
Hicks, B. M., Krueger, R. F., Iacono, W. G., McGue, M. & Patrick, C. J. Family transmission and heritability of externalizing disorders: a twin-family study. Arch. Gen. Psychiatry 61, 922–928 (2004).
Krueger, R. F. et al. Etiologic connections among substance dependence, antisocial behavior and personality: modeling the externalizing spectrum. J. Abnorm. Psychol. 111, 411–424 (2002).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2018).
Swann, A. C., Lijffijt, M., O’Brien, B. & Mathew, S. J. Impulsivity and suicidal behavior. Curr. Top. Behav. Neurosci. 47, 179–195 (2020).
Zhou, H. et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA Psychiatry https://doi.org/10.1001/jamapsychiatry.2020.1206 (2020).
Mullins, N. et al. GWAS of suicide attempt in psychiatric disorders and association with major depression polygenic risk scores. Am. J. Psychiatry 176, 651–660 (2019).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019).
Kendler, K. S. & Myers, J. The boundaries of the internalizing and externalizing genetic spectra in men and women. Psychol. Med. 44, 647–655 (2013).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Walters, R. K. et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 21, 1656–1669 (2018).
Sanchez-Roige, S. et al. Genome-wide association study meta-analysis of the alcohol use disorders identification test in two population-based cohorts. Am. J. Psychiatry 176, 107–118 (2018).
Pasman, J. A. et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat. Neurosci. 21, 1161–1170 (2018).
Karlsson Linnér, R. et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat. Genet. 51, 245–257 (2019).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Lee, P. H. et al. Genomic relationships, novel loci and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482 (2019).
Lo, M.-T. et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat. Genet. 49, 152–156 (2016).
Rosenström, T. et al. Joint factorial structure of psychopathology and personality. Psychol. Med. 49, 2158–2167 (2019).
Townsend, P. Health and Deprivation: Inequality and the North (Croom Helm, 1988).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
de la Fuente, J., Davies, G., Grotzinger, A. D., Tucker-Drob, E. M. & Deary, I. J. A general dimension of genetic sharing across diverse cognitive traits inferred from molecular data. Nat. Hum. Behav. 5, 49–58 (2021).
Hart, A. B. & Kranzler, H. R. Alcohol dependence genetics: lessons learned from genome-wide association studies (GWAS) and post-GWAS analyses. Alcohol. Clin. Exp. Res. 39, 1312–1327 (2015).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, 1–19 (2015).
Sey, N. Y. A. et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 23, 583–593 (2020).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Jaykumar, A. B. et al. Role of Alström syndrome 1 in the regulation of blood pressure and renal function. JCI Insight 3, e95076 (2018).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Li, Y. R. et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 21, 1018–1027 (2015).
Sanchez-Roige, S. et al. Genome-wide association studies of impulsive personality traits (BIS-11 and UPPS-P) and drug experimentation in up to 22,861 adult research participants identify loci in the CACNA1I and CADM2 genes. J. Neurosci. 39, 2562–2572 (2019).
Zhao, B. et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet. 51, 1637–1644 (2019).
Edenberg, H. J. et al. Variations in GABRA2, encoding the α2 subunit of the GABAA receptor, are associated with alcohol dependence and with brain oscillations. Am. J. Hum. Genet. 74, 705–714 (2004).
Dick, D. M. et al. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav. Genet. 36, 577–590 (2006).
Duman, R. S., Sanacora, G. & Krystal, J. H. Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron 102, 75–90 (2019).
Brambilla, P., Perez, J., Barale, F., Schettini, G. & Soares, J. C. GABAergic dysfunction in mood disorders. Mol. Psychiatry 8, 721–737 (2003).
Okbay, A. et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016).
Hill, W. D. et al. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry 24, 169–181 (2019).
Schrode, N. et al. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 51, 1475–1485 (2019).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Derzon, J. H. The correspondence of family features with problem, aggressive, criminal and violent behavior: a meta-analysis. J. Exp. Criminol. https://doi.org/10.1007/s11292-010-9098-0 (2010).
O’Brien, D. T., Farrell, C. & Welsh, B. C. Broken (windows) theory: a meta-analysis of the evidence for the pathways from neighborhood disorder to resident health outcomes and behaviors. Soc. Sci. Med. https://doi.org/10.1016/j.socscimed.2018.11.015 (2019).
Chang, L. Y., Wang, M. Y. & Tsai, P. S. Neighborhood disadvantage and physical aggression in children and adolescents: a systematic review and meta-analysis of multilevel studies. Aggress. Behav. https://doi.org/10.1002/ab.21641 (2016).
Davis, L. Psychiatric genomics, phenomics and ethics research in a 270,000-person Biobank (BioVU). Eur. Neuropsychopharmacol. 29, S739–S740 (2019).
Young, A. I., Benonisdottir, S., Przeworski, M. & Kong, A. Deconstructing the sources of genotype–phenotype associations in humans. Science 365, 1396–1400 (2019).
Kong, A. et al. The nature of nurture: effects of parental genotypes. Science 359, 424–428 (2018).
Selzam, S. et al. Comparing within- and between-family polygenic score prediction. Am. J. Hum. Genet. 105, 351–363 (2019).
Ripke, S. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Tielbeek, J. J. et al. Genome-wide association studies of a broad spectrum of antisocial behavior. JAMA Psychiatry 74, 1242–1250 (2017).
Kranzler, H. R. et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 10, 1499 (2019).
Bulik-Sullivan, B. K. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Anttila, V. et al. Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757 (2018).
Gage, S. H., Smith, G. D., Ware, J. J., Flint, J. & Munafò, M. R. G = E: what GWAS can tell us about the environment. PLoS Genet. 12, e1005765 (2016).
Fox, D. Subversive science. Penn State Law Rev. 124, 153–191 (2019).
American Society of Human Genetics. ASHG denounces attempts to link genetics and racial supremacy. Am. J. Hum. Genet. 103, 636 (2018).
Kvaale, E. P., Gottdiener, W. H. & Haslam, N. Biogenetic explanations and stigma: a meta-analytic review of associations among laypeople. Soc. Sci. Med. 96, 95–103 (2013).
Tucker-Drob, E. M., Briley, D. A. & Harden, K. P. Genetic and environmental influences on cognition across development and context. Curr. Dir. Psychol. Sci. 22, 349–355 (2013).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Yang, J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Walter, K. et al. The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015).
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Rietveld, C. A. et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc. Natl Acad. Sci. USA 111, 13790–13794 (2014).
Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016).
Harris, K. M., Halpern, C. T., Haberstick, B. C. & Smolen, A. The National Longitudinal Study of Adolescent Health (Add Health) sibling pairs data. Twin Res. Hum. Genet. 16, 391–398 (2013).
McQueen, M. B. et al. The National Longitudinal Study of Adolescent to Adult Health (Add Health) sibling pairs genome-wide data. Behav. Genet. 45, 12–23 (2015).
Begleiter, H. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res. World 19, 228–236 (1995).
Edenberg, H. J. The collaborative study on the genetics of alcoholism: an update. Alcohol Res. Health 26, 214–218 (2002).
Bucholz, K. K. et al. Comparison of parent, peer, psychiatric and cannabis use influences across stages of offspring alcohol involvement: evidence from the COGA Prospective Study. Alcohol. Clin. Exp. Res. https://doi.org/10.1111/acer.13293 (2017).
Calkins, M. E. et al. The Philadelphia Neurodevelopmental Cohort: constructing a deep phenotyping collaborative. J. Child Psychol. Psychiatry 56, 1356–1369 (2016).
Satterthwaite, T. D. et al. The Philadelphia Neurodevelopmental Cohort: a publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage 124, 1115–1119 (2016).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Ge, T., Chen, C.-Y., Ni, Y., Feng, Y.-C. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Altshuler, D. M., Gibbs, R. A. & Peltonen, L. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Wei, W.-Q. et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE 12, e0175508 (2017).
Hubbard, T. et al. The Ensembl genome database project. Nucleic Acids Res. 30, 38–41 (2002).
Consortium, T. G. O. The Gene Ontology project in 2008. Nucleic Acids Res. 36, D440–D444 (2007).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464 (2018).
Won, H. et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016).
Rajarajan, P. et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362, eaat4311 (2018).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1–20 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Singhal, A. et al. Multiscale community detection in Cytoscape. PLoS Comput. Biol. 16, e1008239 (2020).
Acknowledgements
This research was carried out under the auspices of the Externalizing Consortium. The study was classified as secondary research of de-identified participants, and the study was awarded ethical approval by the internal review board of Virginia Commonwealth University (VCU), with reference number HM20019386. These analyses were made possible by the generous public sharing of summary statistics from published GWAS from the PGC, the Million Veterans Program, the International Cannabis Consortium, the GWAS & Sequencing Consortium of Alcohol and Nicotine use, the Social Science Genetics Association Consortium, the Genetics of Personality Consortium and the Broad Antisocial Behavior Consortium. We thank the many studies that made these consortia possible, the researchers involved and the participants in those studies, without whom this effort would not be possible. We also thank the research participants and employees of 23andMe for making this work possible. This research was conducted in part using the UKB resource under applications 40830 and 11425. We thank all UKB cohort participants for making this study possible. We thank L. K. Davis for providing access to BioVU. Finally, we thank COGA; principal investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut; and collaborators at eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H. J. Edenberg, J. Nurnberger Jr., T. Foroud and Y. Liu); University of Iowa (S. Kuperman and J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz and A. Agrawal); University of California, San Diego (M. Schuckit); Rutgers University (J. Tischfield and A. Brooks); Department of Biomedical and Health Informatics, The Children’s Hospital of Philadelphia; Department of Genetics, Perelman School of Medicine, University of Pennsylvania (L. Almasy); Virginia Commonwealth University (D.M.D); Icahn School of Medicine at Mount Sinai (A. Goate); and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); J. McClintick, L. Wetherill, X. Xuei, D. Lai, S. O’Connor, M. Plawecki and S. Lourens (Indiana University); G. Chan (University of Iowa and University of Connecticut); J. Meyers, D. Chorlian, C. Kamarajan, A. Pandey and J. Zhang (SUNY Downstate); J. C. Wang, M. Kapoor and S. Bertelsen (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon and S. Saccone (Washington University); J. Salvatore, F. Aliev and B. Cho (Virginia Commonwealth University); and M. Kos (University of Texas Rio Grande Valley). A. Parsian and H. Chen are the National Institute on Alcohol Abuse and Alcoholism (NIAAA) staff collaborators. All studies included in the externalizing GWAS are listed in the Supplementary Information. Funding: The Externalizing Consortium has been supported by the NIAAA through an administrative supplement (R01AA015416) and by the National Institute of Drug Abuse (R01DA050721). D.M.D. was supported through funding from the NIAAA (K02AA018755, U10AA008401 and P50AA022527). P.D.K. was supported through a European Research Council Consolidator Grant (647648 EdGe). K.P.H. was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD; R01HD092548 and R01HD083613) and the Jacobs Foundation. A.A.P. was supported by the NIAAA (R01AA026281) and the National Institute of Drug Abuse (P50DA037844). S.S.-R. was supported through a NARSAD Young Investigator Award from the Brain and Behavior Foundation (grant no. 27676). Both A.A.P. and S.S.-R. were supported by funds from the California Tobacco-Related Disease Research Program (grant nos. 28IR-0070 and T29KT0526). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the above funding bodies. This research used data from Add Health, a program project directed by K.M.H. (principal investigator) and designed by J. R. Udry, P. S. Bearman and K.M.H. at the University of North Carolina at Chapel Hill, and funded by grant P01HD031921 from the Eunice Kennedy Shriver NICHD, with cooperative funding from 23 other federal agencies and foundations. Information on how to obtain the Add Health data files is available on the Add Health website (https://addhealth.cpc.unc.edu/). This research used Add Health GWAS data funded by Eunice Kennedy Shriver NICHD grants R01HD073342 to K.M.H. (principal investigator) and R01HD060726 to K.M.H., J. D. Boardman, and M. B. McQueen (multiple principal investigators). COGA is a national collaborative study supported by the National Institutes of Health (NIH) grant U10AA008401 from the NIAAA and the National Institute on Drug Abuse. Data were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by numerous sources, including institutional funding, private agencies and federal grants. These include the NIH-funded shared instrumentation grant S10RR025141, and CTSA grants UL1TR002243, UL1TR000445 and UL1RR024975. Genomic data are also supported by investigator-led projects, including U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962 and R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu-funding/. Support for data collection for the PNC, acquired through dbGaP (accession no. phs000607, v3.p2), was provided by grant RC2MH089983 awarded to R. Gur and RC2MH089924 was awarded to H. Hakonarson. Participants were recruited and genotyped through the Center for Applied Genomics (CAG) at The Children’s Hospital in Philadelphia (CHOP). Phenotypic data collection occurred at the CAG/CHOP and at the Brain Behavior Laboratory, University of Pennsylvania. A full list of funding for investigator effort is available in the Supplementary Information.
Author information
Authors and Affiliations
Consortia
Contributions
D.M.D. and P.D.K. conceived the study. The study protocol was developed by D.M.D., K.P.H., R.K.L., P.D.K., T.T.M. and A.A.P. D.M.D., K.P.H., P.D.K. and A.A.P. jointly oversaw the study. D.M.D. and R.K.L. led the writing of the manuscript, with substantive contributions to the writing from K.P.H., P.D.K. and A.A.P. R.K.L. and T.T.M. were the lead analysts, responsible for conducting GWAS, quality control, meta-analysis, genetic correlations and multivariate analyses with genomic SEM, with assistance from A.D.G. R.K.L. led the proxy-phenotype and quasi-replication analyses. P.B.B. led the polygenic score analyses, and R.K.L. and T.T.M. contributed to those analyses. S.S.-R. performed the PheWAS in BioVU. R.d.V. derived analytical s.e. for the within-family analyses. S.S.-R. led the bioinformatics analyses, and R.K.L, S.B.R. and T.I. contributed to those analyses. P.B.B., R.K.L, T.T.M. and S.S.-R. prepared the tables and figures, with assistance from M.N.D., J.W.M. and H.E.P. J.J.T., E.C.J., M.L., H.Z., R.K. and J.A.P. prepared cohort-level GWAS meta-analyses under the supervision of K.J.H.V., D.J.L., S.V., H.R.K. and J.G. K.M.H. assisted with analyses performed in the Add Health study cohort. A.D.G., E.M.T.-D. and I.D.W. provided helpful advice and feedback on various aspects of the study design. All authors contributed to and critically reviewed the manuscript. R.K.L., T.T.M., P.B.B. and S.S.-R. made especially major contributions to the writing and editing.
Corresponding authors
Ethics declarations
Competing interests
H.R.K. is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last 3 years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor and Amygdala Neurosciences. H.R.K. and J.G. are named as inventors on PCT patent application no. 15/878,640 entitled ‘genotype-guided dosing of opioid agonists,’ filed on 24 January 2018. J.G. did paid editorial work for the journal Complex Psychiatry. The authors declare no other competing interests.
Additional information
Peer review information Nature Neuroscience thanks Eske Derks and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
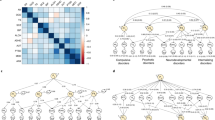
Extended Data Fig. 1 Genetic correlations with the genetic externalizing factor (EXT).
Dot plot of genetic correlations (rg) estimated with Genomic SEM between the genetic externalizing factor (EXT) with 91 other complex traits (Supplementary Methods). Error bars are 95% confidence intervals, calculated as 1.96 × SE, centered on the rg estimate (omitted for Agreeableness). The estimates are also reported in Supplementary Table 8, together with the exact number of independent samples used to derive each estimate. This figure displays genetic correlations with personality measures based on GWAS summary statistics from the Genomics of Personality Consortium, while Fig. 1 instead reports genetic correlations with personality measures based on more recent and substantially larger GWAS provided by 23andMe.
Extended Data Fig. 2 Quantile-quantile (Q-Q) plots of the externalizing GWAS and QSNP results.
The panels display Q-Q plots for (a) the externalizing GWAS (Neff = 1,492,085), and (b) SNP-level tests of heterogeneity (QSNP) with respect to the SNP-effects estimated in the externalizing GWAS (for more details see Supplementary Information section 3). The y-axis is the observed association P value on the –log10 scale (based on a two-sided Z-test in a, and based on a one-sided χ2 test scaled to 1 degree of freedom in b). The gray shaded areas represent 95% confidence intervals centered on the expected –log10(P) of the null distribution. The genomic inflation factors displayed here, λGC, is defined as the median χ2 association test statistic divided by the expected median of the χ2 distribution with 1 degree of freedom, and were calculated with 6,132,068 and 6,107,583 SNPs for (a) and (b), respectively. Although there is a noticeable early ‘lift-off’, the estimated LD Score regression intercepts of (a) 1.115 (SE = 0.019) and (b) 0.9556 (SE = 0.013) suggest that most of the inflation of the test statistics is attributable to polygenicity rather than bias from population stratification.
Extended Data Fig. 3 Quantile-quantile (Q-Q) plots of the proxy-phenotypes analyses.
Panels (a–b) show –log10(P values from a two-sided Z-test) for linear regression of the 553 and 579 EXT SNPs (or such SNPs that could be proxied in case of missingness, r2 > 0.8) that were looked up in independent, second-stage GWAS samples on (1) antisocial behavior (N = 32,574) and (2) alcohol use disorder (N = 202,400), respectively (Supplementary Information section 4). Dashed line denotes experiment-wide significance at P < 0.05/553 and 0.05/579 for (1) and (2), respectively. Enrichment P value is the result of a one-sided test of joint enrichment with the non-parametric Mann-Whitney test against an empirical null distribution of 138,250 and 144,750 near-independent (r2 < 0.1) SNPs, matched on MAF, that were randomly selected from the GWAS on (1) and (2), respectively. Sign concordance is the proportion of looked-up SNPs with concordant direction of effect sizes across the externalizing GWAS and the second-stage GWAS, and the sign concordance P value is from a one-sided binomial tests of the sign concordance for the 579 SNPs (against the null hypothesis of 50% concordance that is expected by chance).
Extended Data Fig. 4 MAGMA gene-based association analysis.
Manhattan plot of the –log10(P from a one-sided Z-test) of 18,093 genes that were tested for association in the MAGMA (v.1.08) gene-based association analysis (Supplementary Information section 6). The 10 most significant genes are labeled with gene names. Red dashed line represents Bonferroni-significance, adjusted for the number of tested genes (one-sided P = 2.74 × 10–6). 928 genes were found to be significant, of which 244 have one or more genome-wide significant SNPs from the externalizing GWAS within their gene breakpoints. The results are also report in Supplementary Table 13.
Extended Data Fig. 5 MAGMA gene-property analysis.
Bar plot of the –log10(P from one-sided Z-tests) of the point estimate from a generalized least squares regression. The analysis identified that the externalizing GWAS is significantly enriched in brain and pituitary gland tissues (Supplementary Information section 6). Dashed line denotes Bonferroni-corrected significance, adjusted for testing 54 tissues (one-sided P < 9.26 × 10–4). 14 tissues were significantly associated with the externalizing GWAS, including 13 brain related tissues and the pituitary tissue. The results are also report in Supplementary Table 15.
Extended Data Fig. 6 MAGMA gene-property analysis of enrichment in brain tissues across 11 developmental stages (BrainSpan).
Bar plot of the –log10(P from one-sided Z-tests) of the point estimate from a generalized least squares regression. The analysis identified that the externalizing GWAS is significantly enriched during prenatal developmental stages (Supplementary Information section 6). Dashed line denotes Bonferroni-corrected significance, adjusted for testing 54 tissues (one-sided P < 9.26 × 10–4). The results are also report in Supplementary Table 16.
Extended Data Fig. 7 Gene overlap across multiple gene-association methods.
Venn diagram illustrating the overlap between (1) the nearest genes to the 579 jointly associated lead SNPs (denoted as the COJO EXT SNPs, see Supplementary Table 9), (2) the genes significant in the MAGMA gene-based analysis (Supplementary Table 13), (3) the genes significant in the H-MAGMA adult brain tissue analysis (Supplementary Table 17), and (4) the genes significant in the S-PrediXcan analysis (Supplementary Table 21). Across these four approaches, 34 genes were consistently implicated; these genes include CADM2, PACSIN3, ZIC4, MAPT, and GABRA2. Colored regions of this diagram correspond to the coloring shown in Supplementary Table 22, which lists all identified genes. No new statistical test was performed to generate this figure, and the statistical test used in each gene-based approach is reported in the notes of Supplementary Tables 9, 13, 17, and 21.
Extended Data Fig. 8 Externalizing systems map estimated with the Order Statistics Local Optimization Method (OSLOM) algorithm.
Representation of the externalizing network neighborhood estimated with PCNet as modular gene systems. In the top panel, circles represent distinct systems, with size indicating the number of genes belonging to each system (min 11 for ‘cilium organization’, and max 379 for the ‘externalizing systems map’). System color indicates the fraction of genes in each system that have been mapped to the externalizing phenotype by at least one of the four gene mapping methods (positional, MAGMA, H-MAGMA, and S-PrediXcan). Systems have been annotated with significantly enriched gene ontology terms. Systems without significant enrichment of biological pathways are labeled with a unique system ID (C454, C461, C453, C462), and may represent novel pathways. (i-vi) Visualization of genes within selected systems that have been mapped to the externalizing phenotype by one or more gene mapping methods, and their molecular interactions. In the bottom panel, the gene size is mapped to the number of methods in which the gene was found associated with externalizing (with the largest genes indicating the gene was identified by all 4 methods), and gene color(s) indicates which method(s) have mapped the gene.
Extended Data Fig. 9 Confirmatory factor analysis of phenotypic externalizing factor in Add Health and COGA.
Path diagram of confirmatory factor analysis (CFA) models in (top panel) Add Health (N = 15,107) and (bottom panel) COGA (N = 16,857) (Supplementary Information section 5). The reported model fit statistics and fit indices are degrees of freedom (df), comparative fit index (CFI), root mean square error (RMSEA), standardized root mean squared residual (SRMR). Standardized factor loadings presented as numbers on the paths.
Supplementary information
Supplementary Information
Supplementary Methods and Supplementary Notes.
Supplementary Data 1
Selected heterogeneity plots.
Supplementary Data 2
Heterogeneity plots for the 579 EXT SNPs.
Rights and permissions
About this article
Cite this article
Karlsson Linnér, R., Mallard, T.T., Barr, P.B. et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci 24, 1367–1376 (2021). https://doi.org/10.1038/s41593-021-00908-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-021-00908-3
This article is cited by
-
Self-control is associated with health-relevant disparities in buccal DNA-methylation measures of biological aging in older adults
Clinical Epigenetics (2024)
-
Genetic factors associated with suicidal behaviors and alcohol use disorders in an American Indian population
Molecular Psychiatry (2024)
-
Examining intergenerational risk factors for conduct problems using polygenic scores in the Norwegian Mother, Father and Child Cohort Study
Molecular Psychiatry (2024)
-
Beyond the factor indeterminacy problem using genome-wide association data
Nature Human Behaviour (2024)
-
Paternal cocaine-seeking motivation defines offspring’s vulnerability to addiction by down-regulating GABAergic GABRG3 in the ventral tegmental area
Translational Psychiatry (2024)