Abstract
Selective neurodegeneration is a critical causal factor in Alzheimer’s disease (AD); however, the mechanisms that lead some neurons to perish, whereas others remain resilient, are unknown. We sought potential drivers of this selective vulnerability using single-nucleus RNA sequencing and discovered that ApoE expression level is a substantial driver of neuronal variability. Strikingly, neuronal expression of ApoE—which has a robust genetic linkage to AD—correlated strongly, on a cell-by-cell basis, with immune response pathways in neurons in the brains of wild-type mice, human ApoE knock-in mice and humans with or without AD. Elimination or over-expression of neuronal ApoE revealed a causal relationship among ApoE expression, neuronal MHC-I expression, tau pathology and neurodegeneration. Functional reduction of MHC-I ameliorated tau pathology in ApoE4-expressing primary neurons and in mouse hippocampi expressing pathological tau. These findings suggest a mechanism linking neuronal ApoE expression to MHC-I expression and, subsequently, to tau pathology and selective neurodegeneration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Mouse snRNA-seq data generated in association with this study are available in the Gene Expression Omnibus under accession number GSE167497. Source data associated with Figs. 4 and 6–8, as well as Extended Data Fig. 9, are available in the Supplementary Information.
snRNA-seq data from the ROSMAP study referenced in Fig. 3, as well as in Extended Data Figs. 5 and 8, are available on the Rush Alzheimer’s Disease Center Research Resource Sharing Hub at https://www.radc.rush.edu/docs/omics.htm (snRNA-seq PFC) or at Synapse (https://www.synapse.org/#!Synapse:syn18485175) under accession number syn18485175. The ROSMAP metadata can be accessed at https://www.synapse.org/#!Synapse:syn3157322.
Data from the Allen Institute for Brain Science, referenced in Extended Data Fig. 6a–d, along with data visualization tools and associated metadata, are available at https://portal.brain-map.org/atlases-and-data/rnaseq.
Single-cell sequencing data from neurotypical human brain, referenced in Extended Data Fig. 6e–g, are available in the Gene Expression Omnibus database: www.ncbi.nlm.nih.gov/geo (accession no. GSE67835).
The Kyoto Encyclopedia of Genes and Genomes Pathways database is available at https://www.genome.jp/kegg/pathway.html. Source data are provided with this paper.
Code availability
All codes generated during this study are available upon reasonable request from the corresponding authors.
References
Fu, H., Hardy, J. & Duff, K. E. Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 21, 1350–1358 (2018).
Andrews-Zwilling, Y. et al. Apolipoprotein E4 causes age- and tau-dependent impairment of GABAergic interneurons, leading to learning and memory deficits in mice. J. Neurosci. 30, 13707–13717 (2010).
Huang, Y. & Mucke, L. Alzheimer mechanisms and therapeutic strategies. Cell 148, 1204–1222 (2012).
Najm, R., Jones, E. A. & Huang, Y. Apolipoprotein E4, inhibitory network dysfunction, and Alzheimer’s disease. Mol. Neurodegener. 14, 24 (2019).
Farrer, L. A. et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. JAMA 278, 1349–1356 (1997).
Ward, A. et al. Prevalence of apolipoprotein E4 genotype and homozygotes (APOE e4/4) among patients diagnosed with Alzheimer’s disease: a systematic review and meta-analysis. Neuroepidemiology 38, 1–17 (2012).
Pitas, R. E., Boyles, J. K., Lee, S. H., Foss, D. & Mahley, R. W. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim. Biophys. Acta 917, 148–161 (1987).
Wang, C. et al. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 24, 647–657 (2018).
Xu, P.-T. et al. Specific regional transcription of apolipoprotein E in human brain neurons. Am. J. Pathol. 154, 601–611 (1999).
Xu, Q. et al. Profile and regulation of apolipoprotein E (apoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the apoE locus. J. Neurosci. 26, 4985–4994 (2006).
Knoferle, J. et al. Apolipoprotein E4 produced in GABAergic interneurons causes learning and memory deficits in mice. J. Neurosci. 34, 14069–14078 (2014).
Brodbeck, J. et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J. Biol. Chem. 286, 17217–17226 (2011).
Lin, Y.-T. et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 98, 1141–1154 (2018).
Shi, Y. et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527 (2017).
Orr, A. L. et al. Neuronal apolipoprotein E4 expression results in proteome-wide alterations and compromises bioenergetic capacity by disrupting mitochondrial function. J. Alzheimers Dis. 68, 991–1011 (2019).
Mahley, R. W., Weisgraber, K. H. & Huang, Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc. Natl Acad. Sci. USA 103, 5644–5651 (2006).
Adelson, J. D. et al. Neuroprotection from stroke in the absence of MHCI or PirB. Neuron 73, 1100–1107 (2012).
Bombeiro, A. L. et al. MHC-I and PirB upregulation in the central and peripheral nervous system following sciatic nerve injury. PLoS ONE 11, e0161463 (2016).
Corriveau, R. A., Huh, G. S. & Shatz, C. J. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron 21, 505–520 (1998).
Mangold, C. A. et al. CNS-wide sexually dimorphic induction of the major histocompatibility complex 1 pathway with aging. J. Gerontol. A Biol. Sci. Med. Sci. 72, 16–29 (2017).
Starkey, H. D. V. et al. Neuroglial expression of the MHCI pathway and PirB receptor is upregulated in the hippocampus with advanced aging. J. Mol. Neurosci. 48, 111–126 (2012).
Datwani, A. et al. Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron 64, 463–470 (2009).
Lee, H. et al. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature 509, 195–200 (2014).
Leung, L. et al. Apolipoprotein E4 causes age- and sex-dependent impairments of hilar GABAergic interneurons and learning and memory deficits in mice. PLoS ONE 7, e53569 (2012).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Cembrowski, M. S., Wang, L., Sugino, K., Shields, B. C. & Spruston, N. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5, e14997 (2016).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030 (2018).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Dijk, Dvan et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729 (2018).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Suberbielle, E. et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 16, 613–621 (2013).
Gerakis, Y. & Hetz, C. A decay of the adaptive capacity of the unfolded protein response exacerbates Alzheimer’s disease. Neurobiol. Aging 63, 162–164 (2018).
Shi, Y. & Holtzman, D. M. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat. Rev. Immunol. 18, 759 (2018).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015).
Ohtsuka, M., Inoko, H., Kulski, J. K. & Yoshimura, S. Major histocompatibility complex (Mhc) class Ib gene duplications, organization and expression patterns in mouse strain C57BL/6. BMC Genomics 9, 178 (2008).
Brecht, W. J. et al. Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 24, 2527–2534 (2004).
Mondragón‐Rodríguez, S., Perry, G., Luna‐Muñoz, J., Acevedo‐Aquino, M. C. & Williams, S. Phosphorylation of tau protein at sites Ser396–404 is one of the earliest events in Alzheimer’s disease and Down syndrome. Neuropathol. Appl. Neurobiol. 40, 121–135 (2014).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Ballatore, C., Lee, V. M.-Y. & Trojanowski, J. Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 (2007).
Germain, R. N. & Margulies, D. H. The biochemistry and cell biology of antigen processing and presentation. Ann. Rev. Immunol. 11, 403–450 (1993).
Hansen, T. H. & Leet, D. R. Mechanism of class I assembly with β2 microglobulin and loading with peptide. In: Advances in Immunology (ed Dixon, F. J.) 64, 105–137 (Academic Press, 1997).
Harris, F. M. et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl Acad. Sci. USA 100, 10966–10971 (2003).
Tesseur, I. et al. Expression of human apolipoprotein E4 in neurons causes hyperphosphorylation of protein tau in the brains of transgenic mice. Am. J. Pathol. 156, 951–964 (2000).
Goddard, C. A., Butts, D. A. & Shatz, C. J. Regulation of CNS synapses by neuronal MHC class I. Proc. Natl Acad. Sci. USA 104, 6828–6833 (2007).
Gate, D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020).
Huh, G. S. et al. Functional requirement for class I MHC in CNS development and plasticity. Science 290, 2155–2159 (2000).
Kim, T. et al. Human LilrB2 is a β-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer’s model. Science 341, 1399–1404 (2013).
Brown, G. C. & Neher, J. J. Microglial phagocytosis of live neurons. Nat. Rev. Neurosci. 15, 209–216 (2014).
Hamanaka, H. et al. Altered cholesterol metabolism in human apolipoprotein E4 knock-in mice. Hum. Mol. Genet. 9, 353–361 (2000).
Bien-Ly, N., Gillespie, A. K., Walker, D., Yoon, S. Y. & Huang, Y. Reducing human apolipoprotein E levels attenuates age-dependent Aβ accumulation in mutant human amyloid precursor protein transgenic mice. J. Neurosci. 32, 4803–4811 (2012).
Bennett, D. A. et al. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 64, S161–S189 (2018).
De Jager, P. L. et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142 (2018).
10x Genomics. Sample Preparation Demonstrated Protocols: Isolation of Nuclei for Single Cell RNA Sequencing https://assets.ctfassets.net/an68im79xiti/6FhJX6yndYy0OwskGmMc8I/e2677be827e82cd954ecfb8b30278e5e/CG000124_SamplePrepDemonstratedProtocol_-_Nuclei_RevD.pdf (2018).
Allen Institute for Brain Science. Allen Cell Types Database Technical White Paper: Transcriptomics. http://help.brain-map.org/download/attachments/8323525/CellTypes_Transcriptomics_Overview.pdf?version=4&modificationDate=1538066831541&api=v2 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, 1–13 (2015). e47.
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
He, Z. et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci. 20, 886–895 (2017).
Acknowledgements
This work was partially supported by grants R01AG048017, RF1AG055421 and R01AG055682 to Y. Huang from the National Institutes of Health (NIH). The results published here are, in part, based on data obtained from the AMP-AD Knowledge Portal accessed at https://doi.org/10.7303/syn2580853. We are grateful to the participants and data contributors in the Religious Order Study, the Memory and Aging Project (ROSMAP). ROSMAP data were provided by the Rush Alzheimer’s Disease Center at Rush University Medical Center. ROSMAP data collection was supported through funding by National Institute on Aging grants P30AG10161, R01AG15819, R01AG17917, R01AG36836, U01AG32984, U01AG46152 and U01061356; the Illinois Department of Public Health; and the Translational Genomics Research Institute. The Gladstone Flow Cytometry Core FACSAria cell sorter is supported by NIH S10 RR028962 and the James B. Pendleton Charitable Trust. We thank E. Chow and the staff at the UCSF Center for Advanced Technology Core for advice and support with RNA sequencing; N. Carli, J. McGuire, P.-L. So and K. Pollard of the Gladstone Genomics Core for advice on sample preparation and sequencing; N. Raman of the Gladstone Flow Cytometry Core for sorting the nuclei; S. Belunek and W. Maguire of Gladstone Information Technology for server support; J. Roudabush and V. Viray for contract support; S. Oduah and L. Hagimori for purchasing; and T. Pak for editorial assistance.
Author information
Authors and Affiliations
Contributions
K.A.Z and Y. Huang designed and coordinated the study and wrote the manuscript. K.A.Z. carried out most studies and data analysis. R.N. conducted immunohistochemical studies. A.L.T. and Y. Hao dissected mouse hippocampi, isolated cell nuclei and prepared samples for RNA sequencing. S.Y.Y. managed mouse lines and contributed to mouse hippocampal virus injection, brain collections, immunohistochemistry and image collection. N.K., M.R.N., A.R., D.J.A., Q.X., A.A. and O.C.T. contributed to biochemical and immunohistochemical studies and data analysis. J.B. contributed to mouse hippocampal virus injection. D.A.B. provided clinical, pathological and snRNA-seq data from human brains of the ROSMAP cohort and critically reviewed the manuscript. Y. Huang supervised the project.
Corresponding authors
Ethics declarations
Competing interests
Y. Huang is a co-founder and scientific advisory board member of E-Scape Bio, GABAeron and Mederon Bio. All other authors declare no competing financial interests.
Additional information
Peer review information Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
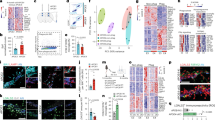
Extended Data Fig. 1 Cell cluster identification and quality control measures in snRNA-seq analysis of apoE-KI mice.
a, Feature plots of imputed expression of marker genes for major cell type clusters, as well as matched whole-brain and hippocampal expression of that marker gene in the Allen Institute for Brain Science Mouse ISH Atlas28. b, Violin plot depicting marker genes for larger cell classes (such as Syn1 for neurons) as well as marker genes for individual clusters, such as C1ql2 for dentate gyrus granule cells, Pdgfra for OPCs, and Folr1 for choroid plexus. c, tSNE plots of all the nuclei broken out by apoE genotype (columns) and mouse age (rows) showing a lack of batch effect by sample and representation of all major cell types in both genotypes at all ages. d, Quality control measures: number of UMIs, number of genes, and percent mitochondrial reads from each cluster.
Extended Data Fig. 2 ApoE correlation with the first two PCs is not driven by age, genotype, cell type markers, or quality control markers.
a–d, PCA plots demonstrating that the correlation between apoE gene expression and the first 2 principal components (PC1 and PC2) across neuronal cell types is not driven by measures of quality control or read depth, such as number of UMIs, number of genes, or percent mitochondrial reads. e–h, Neither is the apoE expression gradient driven by apoE genotype or mouse age. i–l, Additionally, this apoE expression gradient is not explained by differences in cell type marker expression, such as Syn1 for neurons or Aqp4 for astrocytes (i–l), indicating that the apoE-expression-high cells are not misclassified neuron/astrocyte doublets.
Extended Data Fig. 3 ApoE and pathway correlations are highly similar across apoE genotype and age.
a,b, Heatmaps showing apoE and pathway correlation across cell types for the top 10 apoE-correlated pathways for each neuronal subtype, broken out by apoE genotype and mouse age, demonstrating a strong conservation of apoE and pathway relationships across apoE genotypes and ages.
Extended Data Fig. 4 Principal components analysis (PCA) of snRNA-seq data reveals the most prominent sources of cell-by-cell variation within each neuronal type in wildtype (WT) mouse cortex and the top correlates of neuronal apoE expression are enriched for cellular stress and immune response pathways in WT mouse cortical neurons.
a, Clustering using the Seurat package revealed 16 distinct cellular populations in WT mouse cortex where neurons were purposefully enriched34. Marker gene analysis led to the identification of 15 neuronal clusters and one cluster of oligodendrocytes. b, ApoE expression across cell types, demonstrating expression of apoE across neuronal types. c, Heatmap illustrating the correlation between apoE expression and KEGG pathway expression scores for the top 10 apoE expression-correlated pathways from each subset of neurons. d, Network visualization of the proportion of shared genes amongst the pathways represented in c. There are two main modules of inter-related pathways. The blue module is related to neurodegenerative disease and includes the Alzheimer disease, Huntington disease, and Parkinson disease pathways. The orange module, consisting of ten apoE-correlated pathways, relates to immune response. e, In Cluster Ex. 1 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = 0.86, p = 1.7 × 10−284) and PC2 (Pearson’s correlation coefficient; r = 0.47, p = 2.5 × 10−54). f, In Cluster Ex.3 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = −0.84, p = 2.5 × 10−121) and PC2 (Pearson’s correlation coefficient; r = 0.24, p = 8.8 × 10−8). g, In Cluster Ex.5 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = −0.98, p = 5.1 × 10−251) and PC2 (Pearson’s correlation coefficient; r = 0.13, p = 0.01). h, In Cluster Ex. 6 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = 0.44, p = 8.0 × 10−17) and PC2 (Pearson’s correlation coefficient; r = 0.88, p = 4.6 × 10−108). i, In Cluster Ex. 7 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = 0.94, p = 2.1 × 10−143) and PC2 (Pearson’s correlation coefficient; r = −0.32, p = 1.8 × 10−8). j, In Cluster Ex. 8 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = 0.96, p = 3.5 × 10−160). k, In Cluster Ex. 9 cells, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = −0.88, p = 3.2 × 10−90) and PC2 (Pearson’s correlation coefficient; r = −0.43, p = 5.4 × 10−14). l, In SST Interneurons, apoE expression is strongly correlated with PC1 (Pearson’s correlation coefficient; r = −0.51, p = 1.5 ×10−18) and PC2 (Pearson’s correlation coefficient; r = −0.81, p = 8.7 × 10−62).
Extended Data Fig. 5 Principal components analysis (PCA) reveals the most prominent sources of cell-by-cell variation across neuronal types in the ROSMAP dataset.
Across multiple human neuronal cell types, apoE expression levels correlate with the first two PCs. a, In Cluster 4 Excitatory neurons (n = 3574), apoE expression is correlated with PC1 (Pearson’s correlation coefficient; r = 0.33, p = 2 × 10−91) and PC2 (Pearson’s correlation coefficient; r = 0.52, p = 3 × 10−255). b, In Cluster 8 Excitatory neurons (n = 2482), apoE expression is correlated with PC1 (Pearson’s correlation coefficient; r = −0.36, p = 2 × 10−80) and PC2 (Pearson’s correlation coefficient; r = 0.54, p = 1 × 10−191). c, In Cluster 11 Excitatory neurons (n = 1492), apoE expression is correlated with PC1 (Pearson’s correlation coefficient; r = 0.27, p = 7 × 10−26) and PC2 (Pearson’s correlation coefficient; r = −0.48, p = 4 × 10−86). d, In Cluster 7 Inhibitory neurons (n = 2537), apoE expression is correlated with PC1 (Pearson’s correlation coefficient; r = −0.45, p = 2 × 10−127) and PC2 (Pearson’s correlation coefficient; r = −0.12, p = 1 × 10−10). e, In Cluster 12 Inhibitory neurons (n = 1425), apoE expression is correlated with PC1 (Pearson’s correlation coefficient; r = −0.40, p = 4 × 10−55) and PC2 (Pearson’s correlation coefficient; r = 0.67, p = 3 × 10−186). f, In Cluster 15 Inhibitory neurons (n = 897), apoE expression is correlated with PC1 (Pearson’s correlation coefficient; r = 0.54, p = 2 × 10−70) and PC2 (Pearson’s correlation coefficient; r = −0.67, p = 7 × 10−120).
Extended Data Fig. 6 Relationships of neuronal apoE and cellular stress and immune response pathways are replicated in additional human brain snRNA-seq datasets.
a, Clustering of a human brain dataset by cell type (https://portal.brain-map.org/atlases-and-data/rnaseq). b, ApoE expression across cell types, demonstrating expression of apoE across neuronal types. c, Heatmap illustrating the correlation between apoE expression and KEGG pathway expression scores for the top 10 apoE expression-correlated pathways from each subset of neurons. d, Network plot illustrating the proportion of shared genes amongst apoE expression-correlated pathways shared between human and mouse. Edge width represents proportion of shared genes. There are two main modules of inter-related pathways. One (blue) module is related to neurodegenerative disease and includes the Alzheimer disease and Huntington disease. The other (orange) module, consisting of eight apoE-correlated pathways, is related to immune response. e, Clustering of another human dataset by cell type36. f, Heatmap illustrating the correlation between apoE expression and KEGG pathway expression scores for the top 10 apoE expression-correlated pathways from each subset of neurons. g, Network plot illustrating the proportion of shared genes amongst apoE expression-correlated pathways. Edge width represents proportion of shared genes. There are two main modules of inter-related pathways. The larger (green) module is related to cellular metabolism. The other (orange) module, consisting of six apoE-correlated pathways, is related to immune response.
Extended Data Fig. 7 Cell cluster identification and apoE expression in the combined set of apoE-KI and apoE-KI/Syn-Cre data.
a, Feature plots of marker genes for major cell types in the combined apoE-KI and apoE-KI/Syn-Cre cell clustering. b, Histograms of apoE expression levels in the combined apoE-KI and apoE-KI/Syn-Cre cohort, showing that even the low levels of apoE expression measured in apoE-KI neurons are true expression, fully separated from the noise levels in apoE-KI/Syn-Cre neurons.
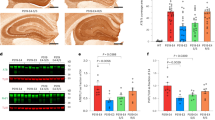
Extended Data Fig. 8 Neuronal expression of apoE predicts neuronal expression of MHC-I genes, and neuronal expression of MHC-I genes predicts Tau tangle, but not β-amyloid, pathology across patients with MCI or AD.
a, Linear regression coefficients (± 95% confidence intervals) for age, sex, apoE4 genotype, clinical diagnosis (MCI or AD relative to control) and average apoE expression level in neurons in predicting the expression of MHC-I genes and B2M gene in neurons of patients from the ROSMAP snRNA-seq cohort depicted in Fig. 3. b, Linear regression coefficients (± 95% confidence intervals) for age, sex, apoE4 genotype, clinical diagnosis (MCI or AD relative to control), and MHC-I genes and B2M gene expression in predicting tau tangle pathology in patients from the ROSMAP snRNA-seq cohort depicted in Fig. 3. c, Linear regression coefficients (± 95% confidence intervals) for age, sex, apoE4 genotype, clinical diagnosis (MCI or AD relative to control), and MHC-I genes and B2M gene expression in predicting β-amyloid pathology in participants from the ROSMAP snRNA-seq cohort depicted in Fig. 3.
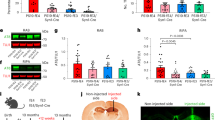
Extended Data Fig. 9 Quantification of B2M protein in B2M-shRNA-treated NSE-E4+/+ mouse primary neurons and B2M-KO mouse primary neurons.
a, Western blot of B2M and TUJ1 protein in NSE-E4+/+ mouse primary neurons treated with Lenti-B2M-shRNA or Lenti-scrambled-shRNA control. Data are representative of two primary neuron culture experiments. b, Quantification of B2M/TUJ1 ratio from western blots. B2M protein is significantly reduced in B2M-shRNA-treated neurons as compared to control shRNA-treated neurons (two-sided t-test, p = 0.017, n = 7 per group). c, Western blot of B2M and TUJ1 protein in lysates of WT and B2M-KO mouse primary neurons, showing elimination of B2M in the B2M-KO neurons. The experiment was performed once.
Extended Data Fig. 10 Model of apoE upregulation of MHC-I driving Tau pathology and selective neuronal and synaptic degeneration/loss.
In response to various cellular stressors during aging, increase in neuronal apoE expression, as a molecular switch, triggers aberrant upregulation of neuronal MHC-I, driving Tau pathology and the selective destruction of individual synapses and neurons, potentially (as a hypothesis) by reactive microglia and/or, MHC’s classical partner, CD8+ T-cells. In the AD context, apoE4 exacerbates this process.
Supplementary information
Supplementary Table 1.
Marker genes for all clusters in all snRNA-seq datasets in Figs. 1, 3 and 6. Includes marker genes for every cluster of cells in the apoE-KI and apoE-KI/Syn-Cre mouse hippocampus as well as in the human MCI/AD prefrontal cortex dataset.
Supplementary Table 2.
Pearson’s correlation coefficient (r) and associated P values for all apoE–pathway correlations represented in Figs. 2 and 3. P values associated with Fig. 4. log fold change and P values associated with Fig. 6h. Includes the statistics associated with the heat maps displayed as Figs. 2a, 3c and 6h and the time courses in Fig. 4e–h.
Supplementary Table 3.
Demographic information related to the human MCI and AD donors represented in Fig. 3. Describes the demographic and basic pathological information of the 48 donors from the ROSMAP cohort included in Fig. 3. AD, Alzheimer’s disease; braaksc, braak score; ceradsc, cerad score; cogdx, cognitive diagnosis; dx, diagnosis; educ, education; gpath, global pathology; mmse, mini-mental state exam; msex, male sex; nft, neurofibrillary tangles (silver stain); tangles, tau tangles (AT8).
Supplementary Table 4.
Demographic information related to the human donors represented in Extended Data Fig. 6a–d. Describes the demographic and basic pathological information from the eight donors in the Allen Brain Atlas Brain Map snRNA-seq data included in Extended Data Fig. 6a–d of this manuscript. C, Caucasian; COD, cause of death; F, female; H, Hispanic; I Iraqi descent; M, male; MTG, middle temporal gyrus; N, neurosurgical; N/A, not applicable; N/Av, not available; P, postmortem; PMI, postmortem interval; RIN, RNA integrity number.
Source data
Source Data Fig. 4
Statistical source data for Fig. 4i,k,l,m.
Source Data Fig. 6
Statistical source data for Fig. 6i,l,k,l,m,n.
Source Data Fig. 6
Western blot source data for Fig. 6i,j.
Source Data Fig. 7
Statistical source data for Fig. 7b,c,d,e,f,h,i,j.
Source Data Fig. 8
Statistical source data for Fig. 8b,c.
Source Data Extended Data Fig. 9
Statistical source data for Extended Data Fig. 9b.
Source Data Extended Data Fig. 9
Western blot source data for Extended Data Fig. 9a,c.
Rights and permissions
About this article
Cite this article
Zalocusky, K.A., Najm, R., Taubes, A.L. et al. Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer’s disease. Nat Neurosci 24, 786–798 (2021). https://doi.org/10.1038/s41593-021-00851-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-021-00851-3
This article is cited by
-
Toward viewing behavior for aerial scene categorization
Cognitive Research: Principles and Implications (2024)
-
Inhibition of asparagine endopeptidase (AEP) effectively treats sporadic Alzheimer’s disease in mice
Neuropsychopharmacology (2024)
-
Cell type-specific roles of APOE4 in Alzheimer disease
Nature Reviews Neuroscience (2024)
-
Tremendous Fidelity of Vitamin D3 in Age-related Neurological Disorders
Molecular Neurobiology (2024)
-
APOE4/4 is linked to damaging lipid droplets in Alzheimer’s disease microglia
Nature (2024)