Abstract
Neurodegeneration occurring in multiple sclerosis (MS) contributes to the progression of disability. It is therefore important to identify and neutralize the mechanisms that promote neurodegeneration in MS. Here, we report that oxidized phosphatidylcholines (OxPCs) found in MS lesions, previously identified as end-product markers of oxidative stress, are potent drivers of neurodegeneration. Cultured neurons and oligodendrocytes were killed by OxPCs, and this was ameliorated by microglia. After OxPC injection, mouse spinal cords developed focal demyelinating lesions with prominent axonal loss. The depletion of microglia that accumulated in OxPC lesions exacerbated neurodegeneration. Single-cell RNA sequencing of lesioned spinal cords identified unique subsets of TREM2high mouse microglia responding to OxPC deposition. TREM2 was detected in human MS lesions, and TREM2−/− mice exhibited worsened OxPC lesions. These results identify OxPCs as potent neurotoxins and suggest that enhancing microglia-mediated OxPC clearance via TREM2 could help prevent neurodegeneration in MS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. There are no restrictions on data availability. The scRNA-seq data comparing cells from the spinal cord of PBS- and PAzePC-injected mice are available at the NCBI Sequence Read Archive with the BioProject accession number PRJNA648663. Mass peaks were identified by experimental m/z searches of the LIPID Metabolites and Pathway Strategies In Silico Structural Database (resource available at https://www.lipidmaps.org/resources/databases/lmissd/search.php).
Code availability
The code for sc-RNAseq analysis can be found in Supplementary Software 1 (scRNA-seq analysis file).
References
Lassmann, H. Multiple sclerosis pathology. Cold Spring Harb. Perspect. Med. https://doi.org/10.1101/cshperspect.a028936 (2018).
Dong, Y. & Yong, V. W. When encephalitogenic T cells collaborate with microglia in multiple sclerosis. Nat. Rev. Neurol. 15, 704–717 (2019).
Rawji, K. S. et al. Deficient surveillance and phagocytic activity of myeloid cells within demyelinated lesions in aging mice visualized by ex vivo live multiphoton imaging. J. Neurosci. 38, 1973–1988 (2018).
Bongarzone, E. R., Pasquini, J. M. & Soto, E. F. Oxidative damage to proteins and lipids of CNS myelin produced by in vitro generated reactive oxygen species. J. Neurosci. Res. 41, 213–221 (1995).
Sun, X. et al. Neutralization of oxidized phospholipids ameliorates non-alcoholic steatohepatitis. Cell Metab. 31, 189–206 (2019).
Que, X. et al. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306 (2018).
Zanoni, I. et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236 (2016).
Di Gioia, M. et al. Endogenous oxidized phospholipids reprogram cellular metabolism and boost hyperinflammation. Nat. Immunol. 21, 42–53 (2020).
Imai, Y. et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133, 235–249 (2008).
Haider, L. et al. Oxidative damage in multiple sclerosis lesions. Brain 134, 1914–1924 (2011).
Kanter, J. L. et al. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nat. Med. 12, 138–143 (2006).
Qin, J., Goswami, R., Balabanov, R. & Dawson, G. Oxidized phosphatidylcholine is a marker for neuroinflammation in multiple sclerosis brain. J. Neurosci. Res. 85, 977–984 (2007).
Prinz, M., Jung, S. & Priller, J. Microglia biology: one century of evolving concepts. Cell 179, 292–311 (2019).
Li, Q. & Barres, B. A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18, 225–242 (2018).
Poliani, P. L. et al. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 125, 2161–2170 (2015).
Wang, Y. M. et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015).
Nugent, A. A. et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron 105, 837–854 (2019).
Coraci, I. S. et al. CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer’s disease brains and can mediate production of reactive oxygen species in response to β-amyloid fibrils. Am. J. Pathol. 160, 101–112 (2002).
Neumann, J. et al. Microglia cells protect neurons by direct engulfment of invading neutrophil granulocytes: a new mechanism of CNS immune privilege. J. Neurosci. 28, 5965–5975 (2008).
Szalay, G. et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat. Commun. 7, 11499 (2016).
Plemel, J. R. et al. Microglia response following acute demyelination is heterogenous and limits infiltrating macrophage dispersion. Sci. Adv. 15, eaay6324 (2020).
Chou, M. Y. et al. Oxidation-specific epitopes are dominant targets of innate natural antibodies in mice and humans. J. Clin. Invest. 119, 1335–1349 (2009).
Palinski, W. et al. Cloning of monoclonal autoantibodies to epitopes of oxidized lipoproteins from apolipoprotein E-deficient mice. Demonstration of epitopes of oxidized low density lipoprotein in human plasma. J. Clin. Invest. 98, 800–814 (1996).
Plemel, J. R. et al. Mechanisms of lysophosphatidylcholine-induced demyelination: a primary lipid disrupting myelinopathy. Glia 66, 327–347 (2018).
Keough, M. B. et al. An inhibitor of chondroitin sulfate proteoglycan synthesis promotes central nervous system remyelination. Nat. Commun. 7, 11312 (2016).
Schliwa, M. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 92, 79–91 (1982).
Oliveira, C. A., Kashman, Y. & Mantovani, B. Effects of latrunculin A on immunological phagocytosis and macrophage spreading-associated changes in the F-actin/G-actin content of the cells. Chem. Biol. Interact. 100, 141–153 (1996).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271 (2019).
Jordao, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019).
Gao, D. et al. Structural basis for the recognition of oxidized phospholipids in oxidized low density lipoproteins by class B scavenger receptors CD36 and SR-BI. J. Biol. Chem. 285, 4447–4454 (2010).
Qin, J. D., Testai, F. D., Dawson, S., Kilkus, J. & Dawson, G. Oxidized phosphatidylcholine formation and action in oligodendrocytes. J. Neurochem. 110, 1388–1399 (2009).
Stemmer, U. et al. Toxicity of oxidized phospholipids in cultured macrophages. Lipids Health Dis. 11, 110 (2012).
Bhatia, T. N. et al. Astrocytes do not forfeit their neuroprotective roles after surviving intense oxidative stress. Front. Mol. Neurosci. 12, 87 (2019).
Loidl, A., Sevcsik, E., Riesenhuber, G., Deigner, H. P. & Hermetter, A. Oxidized phospholipids in minimally modified low density lipoprotein induce apoptotic signaling via activation of acid sphingomyelinase in arterial smooth muscle cells. J. Biol. Chem. 278, 32921–32928 (2003).
Skundric, D. S., Cai, J., Cruikshank, W. W. & Gveric, D. Production of IL-16 correlates with CD4+ Th1 inflammation and phosphorylation of axonal cytoskeleton in multiple sclerosis lesions. J. Neuroinflammation 3, 13 (2006).
Pande, A. H., Kar, S. & Tripathy, R. K. Oxidatively modified fatty acyl chain determines physicochemical properties of aggregates of oxidized phospholipids. Biochim. Biophys. Acta 1798, 442–452 (2010).
Chang, M. K., Binder, C. J., Torzewski, M. & Witztum, J. L. C-reactive protein binds to both oxidized LDL and apoptotic cells through recognition of a common ligand: phosphorylcholine of oxidized phospholipids. Proc. Natl Acad. Sci. USA 99, 13043–13048 (2002).
Bergmark, C. et al. A novel function of lipoprotein [a] as a preferential carrier of oxidized phospholipids in human plasma. J. Lipid Res. 49, 2230–2239 (2008).
Leibundgut, G. et al. Determinants of binding of oxidized phospholipids on apolipoprotein (a) and lipoprotein (a). J. Lipid Res. 54, 2815–2830 (2013).
Palavra, F. et al. New markers of early cardiovascular risk in multiple sclerosis patients: oxidized-LDL correlates with clinical staging. Dis. Markers 34, 341–348 (2013).
Soilu-Hanninen, M. et al. High sensitivity measurement of CRP and disease progression in multiple sclerosis. Neurology 65, 153–155 (2005).
Romero, F. et al. A pneumocyte–macrophage paracrine lipid axis drives the lung toward fibrosis. Am. J. Respir. Cell Mol. Biol. 53, 74–86 (2015).
Bieghs, V. et al. Trapping of oxidized LDL in lysosomes of Kupffer cells is a trigger for hepatic inflammation. Liver Int. 33, 1056–1061 (2013).
Benedek, G. et al. MIF and D-DT are potential disease severity modifiers in male MS subjects. Proc. Natl Acad. Sci. USA 114, E8421–E8429 (2017).
van der Poel, M. et al. Transcriptional profiling of human microglia reveals grey–white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun. 10, 1139 (2019).
Andersson, A. et al. Pivotal advance: HMGB1 expression in active lesions of human and experimental multiple sclerosis. J. Leukoc. Biol. 84, 1248–1255 (2008).
Vercellino, M. et al. Progranulin expression in brain tissue and cerebrospinal fluid levels in multiple sclerosis. Mult. Scler. 17, 1194–1201 (2011).
Ma, J. et al. Microglial cystatin F expression is a sensitive indicator for ongoing demyelination with concurrent remyelination. J. Neurosci. Res. 89, 639–649 (2011).
Takahashi, K., Prinz, M., Stagi, M., Chechneva, O. & Neumann, H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 4, e124 (2007).
Polman, C. H. et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 69, 292–302 (2011).
Dhaeze, T. et al. CD70 defines a subset of proinflammatory and CNS-pathogenic TH1/TH17 lymphocytes and is overexpressed in multiple sclerosis. Cell. Mol. Immunol. 16, 652–665 (2019).
Kuhlmann, T. et al. An updated histological classification system for multiple sclerosis lesions. Acta. Neuropathol. 133, 13–24 (2017).
Caughlin, S., Park, D. H., Yeung, K. K., Cechetto, D. F. & Whitehead, S. N. Sublimation of DAN matrix for the detection and visualization of gangliosides in rat brain tissue for MALDI imaging mass spectrometry. J. Vis. Exp. 121, 55254 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Sloka, S., Zhornitsky, S., Silva, C., Metz, L. M. & Yong, V. W. 1,25-Dihydroxyvitamin D3 protects against immune-mediated killing of neurons in culture and in experimental autoimmune encephalomyelitis. PLoS ONE 10, e0144084 (2015).
Mishra, M. K. et al. Laquinimod reduces neuroaxonal injury through inhibiting microglial activation. Ann. Clin. Transl Neurol. 1, 409–422 (2014).
Cua, R. C. et al. Overcoming neurite-inhibitory chondroitin sulfate proteoglycans in the astrocyte matrix. Glia 61, 972–984 (2013).
Lau, L. W. et al. Chondroitin sulfate proteoglycans in demyelinated lesions impair remyelination. Ann. Neurol. 72, 419–432 (2012).
Acknowledgements
We thank C. Silva, R. Chan, the Hotchkiss Brain Institute Advanced Microscopy Platform Facility and the Centre for Health Genomics and Informatics Bioinformatics Core, University of Calgary, for technical help. The authors’ research is supported by operating grants from the Alberta Innovates Health Solutions CRIO Team program, the MS Society of Canada and the Canadian Institutes of Health Research. Y.D. acknowledges postdoctoral fellowship support from the Canadian Institutes of Health Research and Alberta MS Collaboration. S.G. acknowledges postdoctoral fellowship support from the Harley N. Hotchkiss Postdoctoral Fellowship. D.K.K. received postdoctoral funding from Alberta Innovates—Health Solutions, the Multiple Sclerosis Society of Canada (MSSOC) and the Eyes High Scholars program of the University of Calgary. B.M.L. and D.B. gratefully acknowledge studentships from the Alberta Graduate Excellence Scholarship and the Canadian Institutes of Health Research (CIHR) Canada Graduate Scholarships, respectively. W.P. acknowledges CGS-M scholarship support from CIHR. S.Z. holds a joint fellowship from the Fonds de Recherche en Santé du Québec and the MSSOC. This work was funded by operating grants from the MSSOC and CIHR to V.W.Y. (grant number 3527 and FDN 167270, respectively) and A.P. (grant number 3188 and PJT166056, respectively). S.N.W. acknowledges funding support for this research from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-04742) and Canada First Research Excellence Fund BrainsCAN Accelerator Award (44531). V.W.Y. and A.P. acknowledge salary support from the Canada Research Chair (Tier 1) program.
Author information
Authors and Affiliations
Contributions
Y.D. conceived the project, designed, performed, analyzed experiments and wrote the first draft of the paper. C.D. was key for the scRNA-seq experiments and provided critical support for the related data analysis. W.P. and S.N.W. were critical for performing IMS. B.M.L. and F.C.M. performed surgeries. D.K.K. provided support and data for MS brain tissues and provided tissue for EAE experiments. S.G. provided aid for the microglia-depletion experiments and provided tissue for the EAE experiments. B.M.L., D.M., D.B., F.C.M. and T.V. also aided in experiments. S.Z. and A.P. provided MS brain specimens for IMS. V.W.Y. helped conceive the project, provided support and experimental design, supervised the overall study and critically edited the manuscript. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Workflow and additional IMS analysis of MS brains.
a, Workflow of immunofluorescence confocal image analysis. b, Representative Luxol fast blue and hematoxylin and eosin histological stain showing NAWM and white matter (WM) lesion from a second MS brain tissue section (left) and MALD-TOF-IMS positive ion mode scan of adjacent tissue section comparing relative levels of [PON-PC + H]+ in the NAWM and WM lesion (right). c, Representative immunofluorescence confocal microscopy images of the adjacent tissue section as in (b), comparing field of views within the NAWM and the WM lesion. Tissue was labeled with DAPI, E06, and CD45. Data representative of 2 MS patients.
Extended Data Fig. 2 EAE and additional cell culture experiments.
a,b, Representative confocal microscopy images comparing NAWM (no hypercellularity regions from peak EAE tissue) and peak and chronic EAE lesions in the mouse spinal cord parenchyma (a) and cerebellar meninges (b). Tissues were labeled with DAPI, E06, and IBA1. c,d, Quantification comparing the number of E06+ particles in the NAWM white matter and lesions of spinal cord (c) and cerebellum (d) of EAE mice (n = 11 animals for NAWM, 8 for peak, 7 for chronic in the spinal cord; n = 5 for NAWM, 6 for peak, 6 for chronic in the cerebellum). Data acquired from 5 to 8 mice over 2 separate experiments. Significance indicated as * p < 0.05, *** p < 0.001, two-tailed paired t-test comparing the lesion and NAWM of each EAE sample. e, Representative confocal microscopy images comparing a FOV within the NAWM with a FOV within an active lesion, labeled with DAPI, E06, and CD68. Image representative of 6 individual MS brain samples. f, Molecular structures of the phospholipid species used in this study. g, Representative widefield microscopy images of primary mouse neurons labeled with Tuj1 24 h after EtOH control, cholesteryl-d7 palmitate (Choles, 60 µg/ml), or oxidized cholesteryl-d7 linoleate (OxCholes, 60 µg/ml) treatments. h, Bar graph comparing the fold change in the number of DAPI+ Tuj1+ primary mouse neurons 24 h after treatments against the EtOH control. i, Representative widefield microscopy images of primary mouse astrocytes labeled with GFAP 24 h after EtOH control, DPPC (50 µM), or POVPC (50 µM) treatments. DAPI was used to label nuclei. j, Bar graph comparing the fold change in the number of DAPI+ GFAP+ primary mouse astrocytes 24 h after treatments against the EtOH control. Data acquired from 2 separate experiments each with 4 replicates. One-way ANOVA comparing the treatments against the EtOH control. All data presented as mean values with error bars showing +/− SD.
Extended Data Fig. 3 Additional spinal cord injection analysis.
a, Representative confocal image showing DAPI, E06, GFAP and IBA1 in the ventral spinal cord white matter 3 days following PBS sham injection. b, Representative widefield light microscopy images of eriochrome cyanine and neutral red labeling of the spinal cord 7 days after PAzePC injection; the lesion is traced by the dashed line. c, Representative confocal microscopy images of PAzePC injected spinal cord white matter after 7 days. Sections were labeled with DAPI, Olig2, MBP and NF-H. d, Histogram and bar graph comparing the distribution of GFAP mean fluorescence intensity (MFI) across 200 µm in length of NAWM or day 7 POVPC lesion in the spinal cord (n = 3 animals for NAWM, 6 for D7; 664 pixels were analyzed per animal). Significance indicated as * p < 0.05, ** p < 0.01, *** p < 0.001, two-tailed, unpaired t-test. e, Representative confocal microscopy images of PAzePC injected spinal cord white matter after 7 days. Sections were labeled with DAPI, E06, GFAP, and. f, Representative confocal images of LPC injected spinal cord white matter after 7 days. Sections were labeled with DAPI, E06, Tuj1 and IBA1. PAzePC and LPC images (a-c, e-f) representative of 2 separate experiments with 3 mice in each experiment. All data presented as mean values with error bars showing +/− SD.
Extended Data Fig. 4 Additional E06 analysis and OxPC neutralization.
a, Representative confocal microscopy images comparing Ctrl PBS injected spinal cord white matter and POVPC injected spinal cord white matter after 1, 3, or 7 days. Sections were labeled with DAPI, E06, Tuj1 and IBA1. Data for each time point was acquired from 2 to 3 separate experiments with 3 to 5 mice in each experiment. b, Representative widefield microscopy images of primary mouse neurons labeled with Tuj1 for tubulin β3 24 h after EtOH control, POVPC (25 µM), POVPC + IgM (10 µg/ml), or POVPC + E06 (10 µg/ml) treatments. DAPI was used to label nuclei. c, Bar graph comparing the fold change in the number of DAPI+ Tuj1+ primary mouse neurons 24 h after treatment, normalized to the EtOH control. Data acquired from 2 separate experiments each with 4 replicates. Significance indicated as *** p < 0.001, one-way ANOVA comparing the treatments against the EtOH control. d and g, Representative confocal images of the ventral spinal cord 3 days after ATP (30 mM) + IgM (500 µg/ml) or ATP (30 mM) + E06 (500 µg/ml) injection. Sections were labeled with DAPI, E06, and IBA1 in d, or with MBP and NF-H in g. e,f, Bar graphs comparing the percent E06+ deposition (e) or percent IBA1+ (f) found per field of view (FOV) in spinal cord of mice 3 days ATP + IgM or ATP + E06 injection. h, Representative particle analysis masks comparing the number of NF-H + axons with surface area smaller or larger than 30 µm2 in the two treatment groups. Data was acquired from 2 separate experiments with 5 mice per experiment. Significance indicated as * p < 0.05, ** p < 0.01, two-tailed, unpaired t-test. All data presented as mean values with error bars showing +/− SD.
Extended Data Fig. 5 Additional neuron microglia co-culture experiments.
a, Representative microscopy images of primary mouse neurons cultured with or without microglia (MG) and then treated with EtOH, POVPC (50 µM), or POVPC with CytoD for 24 h, labeled with DAPI, Tuj1, CD68, and E06. b, Representative images of mouse neurons treated with 0.015% EtOH control (top left), pre-treatment with 200 nM latrunculin A (LA) for 30 min followed by 0.015% EtOH (top middle) or 50 µM POVPC (top right); and neurons and microglia pre-treated with LA for 30 min followed by EtOH (bottom left), POVPC (bottom middle), or LA for 30 min followed by 50 µM POVPC treatment for 24 h (bottom right). c, Bar graph comparing the fold change in the number of DAPI+ Tuj1+ cells after treatments with or without POVPC, LA, and microglia. Data acquired from 2 separate experiments each with 4 replicates. Significance indicated as * p < 0.05, *** p < 0.001, one-way ANOVA comparing the treatments against the EtOH control. All data presented as mean values with error bars showing +/− SD.
Extended Data Fig. 6 Microglia depletion for days 7–14 after spinal cord OxPC injection.
a, Representative confocal microscopy of uninjured spinal cords isolated from tamoxifen treated CX3CR1creER:Rosa26iDTR mice injected with PBS or DT once every other day, for 7 days. Sections were labeled with DAPI, E06, GFAP, IBA1 on the top panel, or with DAPI, Olig2, MBP, and NF-H on the bottom. b, Schematic of experimental design for day 7–14 microglia depletion. c, Representative confocal microscopy of POVPC induced lesions in the spinal cords from PBS or DT injected mice labeled with DAPI, CD68, and Tmem119. d, Representative confocal microscopy of POVPC induced lesions in the spinal cords from PBS or DT injected mice labeled with DAPI, E06, GFAP and IBA1. e, Representative immunofluorescent laser confocal microscopy of POVPC induced lesions in the spinal cords from PBS or DT injected mice labeled with DAPI, Olig2, MBP and NF-H. Dotted line indicate the lesion ROI selected for image analysis. ROI for the spinal cord lesion from PBS or DT treated mice was determined using CD68 (c, g), IBA1 (d, f, h, j), MBP (e, k, l). f, Bar graph comparing the size of POVPC induced lesion marked by IBA1 accumulation PBS Ctrl and DT treated mice. g, Bar graph comparing the proportion of CD68+ cells and Tmem119+ cells in the POVPC induced lesion from mice treated with PBS or DT. h-m, Bar graphs comparing the size of E06+ area in the ROI (h), percent of the ROI that is E06+ (i), percent of the ROI that is GFAP+ (j), the number of Olig2+ cells per mm2 in the ROI (k), the total number of MBP+ particles in the ROI (l), and the number of NF-H+ axons per mm2 in the ROI (m) between the PBS or DT treated mice 7 days after POVPC injection (n = 7 animals for POVPC, 9 for POVPC + DT). Significance indicated as * p < 0.05, ** p < 0.01, *** p < 0.001, two tailed non-paired t-test. All data presented as mean values with error bars showing +/− SD.
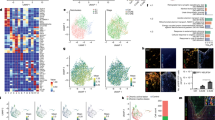
Extended Data Fig. 7 Additional ScRNAseq analysis.
a, Graphs show the distribution of cells from scRNAseq by the number of total unique genes (left), by the number of UMIs (middle), and by percent mitochondrial content (right). b, Graph shows the top 2000 variable features with the top 10 labeled. c, tSNE plot of 5486 cells from sham or OxPC lesion carrying spinal cords displays the separation of cells into 16 clusters (right plot). Left plot displays cells by their origin, PBS injected sham mice (green) or PAzePC injected lesion mice (red). d, Heatmap showing the top 10 differentially expressed genes for clusters. Differentially expressed genes were determined using a t-test and comparison was made for cells in each respective cluster vs. cells in all other clusters Yellow indicates upregulation, purple indicates downregulation.
Extended Data Fig. 8 IPA analysis for homeostatic MGs.
Ingenuity pathway analysis of homeostatic MG subsets from scRNAseq analysis. The top Regulator Effect Network showing cell function changes from each cluster are illustrated for (a) Homeostatic MG1, (b) Homeostatic MG2, and (c) Homeostatic MG3 clusters. P-values are displayed below gene names.
Extended Data Fig. 9 IPA analysis for activated MG/Macs.
Ingenuity pathway analysis of activated MG/Mac subsets from scRNAseq analysis. The top Regulator Effect Network showing cell function changes from each cluster are illustrated for (a) Activated MG/Mac1, (b) Activated MG/Mac2, (c) Activated MG/Mac3, (d) Activated MG/Mac4, (e) Activated MG/Mac5, and (f) Activated MG/Mac6 clusters. P-values are displayed below gene names.
Extended Data Fig. 10 Additional in vitro and in vivo TREM2 experiments.
a, Representative image showing CD36 and b, TREM2 expression in the spinal cord white matter lesion (n = 11 animals for ctrl, 8 for D1, 6 for D3, 9 for D7). Significance indicated as ***p < 0.001, one-way ANOVA comparing all timepoints against the ctrl. c, Representative images showing DAPI (blue), E06 (green), CD45 (red), and TREM2 (cyan) in the NAWM or lesion of MS brain samples. Data representative of 6 individual MS brain samples. d, Graph showing relative fluorescence in ELISA detecting the binding of bovine serum albumin (BSA), E06 antibody, and recombinant mouse TREM2 protein to OxPC at various concentrations. Data shown representative of 3 experiments with 2 replicates per experiment. e, Representative widefield microscopy images of microglia isolated from the adult brains of TREM2+/+ or TREM2−/− mice, or microglia isolated from mix glia cultures grown from neonatal mouse brain isolates. Cells were labeled with DAPI (blue), IBA1 (green) and CD68 (red). f, Bar graph comparing the fold change in the number of DAPI+ CD68+ TREM2+/+ or TREM2−/− microglia 24 h after EtOH or POVPC treatment. Data acquired from 2 separate experiments each with 4 replicates. ANOVA compared the treatments against TREM2+/+ microglia with EtOH control. All data presented as mean values with error bars showing +/− SD.
Supplementary information
Supplementary Table 1
Positive differentially expressed genes from each cluster identified from scRNA-seq. Two-sided Wilcoxon rank-sum test with default wilcox.test settings in R was used to determine significance.
Supplementary Table 2
Statistical summary of exact n values, P values and statistical tests.
Supplementary Software 1
R script code used for scRNA-seq analysis.
Supplementary Video 1
Live imaging of mouse neurons treated with control concentrations of ethanol over 4 h. Neurons were labeled with calcein AM, Hoechst dye and PI. Representative of eight replicates over two independent experiments.
Supplementary Video 2
Live imaging of mouse neurons treated with 100 µM of DPPC over 4 h. Neurons were labeled with calcein AM, Hoechst dye and PI. Representative of eight replicates over two independent experiments.
Supplementary Video 3
Live imaging of mouse neurons treated with 100 µM of POVPC over 4 h. Neurons were labeled with calcein AM, Hoechst dye and PI. Representative of eight replicates over two independent experiments.
Supplementary Video 4
Live imaging of mouse neurons treated with 100 µM of PAzePC over 4 h. Neurons were labeled with calcein AM, Hoechst dye and PI. Representative of eight replicates over two independent experiments.
Supplementary Video 5
Live imaging of mouse neurons treated with 100 µM of LPC over 4 h. Neurons were labeled with calcein AM, Hoechst dye and PI. Representative of eight replicates over two independent experiments.
Rights and permissions
About this article
Cite this article
Dong, Y., D’Mello, C., Pinsky, W. et al. Oxidized phosphatidylcholines found in multiple sclerosis lesions mediate neurodegeneration and are neutralized by microglia. Nat Neurosci 24, 489–503 (2021). https://doi.org/10.1038/s41593-021-00801-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-021-00801-z
This article is cited by
-
LXR agonism for CNS diseases: promises and challenges
Journal of Neuroinflammation (2024)
-
Microglia regulation of central nervous system myelin health and regeneration
Nature Reviews Immunology (2024)
-
Type I Interferon Signalling and Ischemic Stroke: Mechanisms and Therapeutic Potentials
Translational Stroke Research (2024)
-
Novel CH25H+ and OASL+ microglia subclusters play distinct roles in cerebral ischemic stroke
Journal of Neuroinflammation (2023)
-
Lipid in microglial biology — from material to mediator
Inflammation and Regeneration (2023)