Abstract
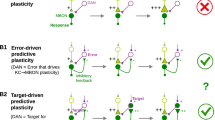
Dopaminergic neurons (DANs) drive learning across the animal kingdom, but the upstream circuits that regulate their activity and thereby learning remain poorly understood. We provide a synaptic-resolution connectome of the circuitry upstream of all DANs in a learning center, the mushroom body of Drosophila larva. We discover afferent sensory pathways and a large population of neurons that provide feedback from mushroom body output neurons and link distinct memory systems (aversive and appetitive). We combine this with functional studies of DANs and their presynaptic partners and with comprehensive circuit modeling. We find that DANs compare convergent feedback from aversive and appetitive systems, which enables the computation of integrated predictions that may improve future learning. Computational modeling reveals that the discovered feedback motifs increase model flexibility and performance on learning tasks. Our study provides the most detailed view to date of biological circuit motifs that support associative learning.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The source data and statistics for Fig. 1 are available in Source Data Fig. 1. The EM volume of the whole CNS26 and the reconstructed neurons and synapses included in this paper are publicly available at https://l1em.catmaid.virtualflybrain.org/, accessible online thanks to the software CATMAID65,66, the same software used to reconstruct and analyze the neuronal circuits included here. Supplementary Adjacency Matrices 1, 2 and 3 contain all the connectivity information for all neurons from the extended MB network discussed in the study. The light microscopy image stacks of genetic driver lines are available from the corresponding authors upon reasonable request.
Code availability
Python code (used for the model in Fig. 7) and Matlab code (used for Supplementary Fig. 3b) can be found in Supplementary Software. The Matlab code for the analysis of imaging and behavioral data is available from the corresponding author upon reasonable request.
References
Schultz, W. Neuronal reward and decision signals: from theories to data. Physiol. Rev. 95, 853–951 (2015).
Dunsmoor, J. E., Niv, Y., Daw, N. & Phelps, E. A. Rethinking extinction. Neuron 88, 47–63 (2015).
Cognigni, P., Felsenberg, J. & Waddell, S. Do the right thing: neural network mechanisms of memory formation, expression and update in drosophila. Curr. Opin. Neurobiol. 49, 51–58 (2018).
Watabe-Uchida, M., Eshel, N. & Uchida, N. Neural circuitry of reward prediction error. Annu. Rev. Neurosci. 40, 373–394 (2017).
Menzel, R. Searching for the memory trace in a mini-brain, the honeybee. Learn. Mem. 8, 53–62 (2001).
Heisenberg, M. Mushroom body memoir: from maps to models. Nat. Rev. Neurosci. 4, 266–275 (2003).
Rescorla, R.A. & Wagner, A.R. in Classical Conditioning II: Current Research and Theory (eds Black, A.H. & Prokasy, W.F.) 64–99 (Appleton Century Crofts, 1972).
Sutton, R. S. & Barto, A. G. Reinforcement Learning: An Introduction (MIT Press, 1998).
Watabe-Uchida, M. The basal ganglia sensory system listens to prefrontal task needs. Neuron 103, 353–355 (2019).
Riemensperger, T., Vller, T., Stock, P., Buchner, E. & Fiala, A. Punishment prediction by dopaminergic neurons in Drosophila. Curr. Biol. 15, 1953–1960 (2005).
Dylla, K. V., Raiser, G., Galizia, C. G. & Szyszka, P. Trace conditioning in Drosophila induces associative plasticity in mushroom body kenyon cells and dopaminergic neurons. Front. Neural Circuits 11, 42 (2017).
Eichler, K. et al. The complete connectome of a learning and memory centre in an insect brain. Nature 548, 175–182 (2017).
Takemura, S. Y. et al. A connectome of a learning and memory center in the adult Drosophila brain. eLife 6, e26975 (2017).
Davis, R. L. Traces of Drosophila memory. Neuron 70, 8–19 (2011).
Thum, A. S. & Gerber, B. Connectomics and function of a memory network: the mushroom body of larval Drosophila. Curr. Opin. Neurobiol. 54, 146–154 (2019).
Handler, A. et al. Distinct dopamine receptor pathways underlie the temporal sensitivity of associative learning. Cell 178, 60–75.e19 (2019).
Hige, T., Aso, Y., Modi, M. N., Rubin, G. M. & Turner, G. C. Heterosynaptic plasticity underlies aversive olfactory learning in Drosophila. Neuron 88, 985–998 (2015).
Waddell, S. Reinforcement signalling in Drosophila; dopamine does it all after all. Curr. Opin. Neurobiol. 23, 324–329 (2013).
Aso, Y. & Rubin, G. M. Dopaminergic neurons write and update memories with cell-type-specific rules. eLife 5, e16135 (2016).
Liu, C. et al. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516 (2012).
Schroll, C. et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr. Biol. 16, 1741–1747 (2006).
Saumweber, T. et al. Functional architecture of reward learning in mushroom body extrinsic neurons of larval Drosophila. Nat. Commun. 9, 1104 (2018).
Caldwell, J. C., Miller, M. M., Wing, S., Soll, D. R. & Eberl, D. F. Dynamic analysis of larval locomotion in Drosophila chordotonal organ mutants. Proc. Natl Acad. Sci. USA 100, 16053–16058 (2003).
Tracey, W., Wilson, R., Laurent, G. & Benzer, S. painless, a Drosophila gene essential for nociception. Cell 113, 261–273 (2003).
Ohyama, T. et al. High-throughput analysis of stimulus-evoked behaviors in Drosophila larva reveals multiple modality-specific escape strategies. PLoS One 8, e71706 (2013).
Ohyama, T. et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639 (2015).
Jovanic, T. et al. Competitive disinhibition mediates behavioral choice and sequences in Drosophila. Cell 167, 858–870.e19 (2016).
Eschbach, C. et al. Associative learning between odorants and mecha nosensory punishment in larval Drosophila. J. Exp. Biol. 214, 3897–3905 (2011).
Liu, W. W. & Wilson, R. I. Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc. Natl Acad. Sci. USA 110, 10294–10299 (2013).
Fushiki, A. et al. A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. eLife 5, e13253 (2016).
Jiang, L. & Litwin-Kumar, A. Models of heterogeneous dopamine signaling in an insect learning and memory center. Preprint at bioRxiv https://doi.org/10.1101/737064 (2019).
Mancini, N. et al. Reversal learning in Drosophila larvae. Learn. Mem. 26, 424–435 (2019).
Felsenberg, J. et al. Integration of parallel opposing memories underlies memory extinction. Cell 175, 709–722.e15 (2018).
Tabone, C. J. & de Belle, J. S. Second-order conditioning in Drosophila. Learn. Mem. 18, 250–253 (2011).
Brembs, B. & Wiener, J. Context and occasion setting in Drosophila visual learning. Learn. Mem. 13, 618–628 (2006).
Schultz, W., Dayan, P. & Montague, P. R. A neural substrate of prediction and reward. Science 275, 1593–1599 (1997).
Zhao, X., Lenek, D., Dag, U., Dickson, B. J. & Keleman, K. Persistent activity in a recurrent circuit underlies courtship memory in Drosophila. eLife 7, e31425 (2018).
Cohn, R., Morantte, I. & Ruta, V. Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell 163, 1742–1755 (2015).
Ichinose, T. et al. Reward signal in a recurrent circuit drives appetitive long-term memory formation. eLife 4, 1–18 (2015).
Ueoka, Y., Hiroi, M., Abe, T. & Tabata, T. Suppression of a single pair of mushroom body output neurons in Drosophila triggers aversive associations. FEBS Open Bio. 7, 562–576 (2017).
Felsenberg, J., Barnstedt, O., Cognigni, P., Lin, S. & Waddell, S. Re-evaluation of learned information in Drosophila. Nature 544, 240–244 (2017).
Pavlowsky, A., Schor, J., Placais, P. Y. & Preat, T. A gabaergic feedback shapes dopaminergic input on the Drosophila mushroom body to promote appetitive long-term memory. Curr. Biol. 28, 1783.e4–1793.e4 (2018).
König, C., Khalili, A., Niewalda, T., Gao, S. & Gerber, B. An optogenetic analogue of second-order reinforcement in Drosophila. Biol. Lett. 15, 20190084 (2019).
Lyutova, R. et al. Reward signaling in a recurrent circuit of dopaminergic neurons and peptidergic kenyon cells. Nat. Commun. 10, 3097 (2019).
Tian, J. et al. Distributed and mixed information in monosynaptic inputs to dopamine neurons. Neuron 91, 1374–1389 (2016).
Beier, K. T. et al. Circuit architecture of vta dopamine neurons revealed by systematic input-output mapping. Cell 162, 622–634 (2015).
Haber, S. N., Fudge, J. L. & McFarland, N. R. Striatonigrostriatal pathways in primates form an ascending spiral from the shell to the dorsolateral striatum. J. Neurosci. 20, 2369–2382 (2000).
Watabe-Uchida, M., Zhu, L., Ogawa, S. K., Vamanrao, A. & Uchida, N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74, 858–873 (2012).
Menegas, W. et al. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife 4, e10032 (2015).
Pfeiffer, B. D. et al. Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Luan, H., Peabody, N. C., Vinson, C. R. & White, B. H. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436 (2006).
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012).
Aso, Y. et al. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577 (2014).
Vogelstein, J. T. et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science 344, 386–392 (2014).
Yang, C. H. et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61, 519–526 (2009).
Awasaki, T., Huang, Y., O’Connor, M. B. & Lee, T. Glia instruct developmental neuronal remodeling through tgf-beta signaling. Nat. Neurosci. 14, 821–823 (2011).
Liu, C. et al. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516 (2012).
Pfeiffer, B. D., Truman, J. W. & Rubin, G. M. Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl Acad. Sci. USA 109, 6626–6631 (2012).
Rohwedder, A. et al. Four individually identified paired dopamine neurons signal reward in larval Drosophila. Curr. Biol. 26, 661–669 (2016).
Suloway, C. et al. Automated molecular microscopy: the new leginon system. J. Struct. Biol. 151, 41–60 (2005).
Cardona, A. et al. Trakem2 software for neural circuit reconstruction. PLoS ONE 7, e38011 (2012).
Saalfeld, S., Fetter, R., Cardona, A. & Tomancak, P. Elastic volume reconstruction from series of ultra-thin microscopy sections. Nat. Methods 9, 717–720 (2012).
Saalfeld, S., Cardona, A., Hartenstein, V. & Tomancak, P. CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984–1986 (2009).
Schneider-Mizell, C. M. et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife 5, e12059 (2016).
Berck, M. E. et al. The wiring diagram of a glomerular olfactory system. eLife 5, e14859 (2016).
Goodman, C. S., Bate, M. & Spitzer, N. C. Embryonic development of identified neurons: origin and transformation of the h cell. J. Neurosci. 1, 94–102 (1981).
Bate, M., Goodman, C. S. & Spitzer, N. C. Embryonic development of identified neurons: segment-specific differences in the h cell homologues. J. Neurosci. 1, 103–106 (1981).
Costa, M., Manton, J. D., Ostrovsky, A. D., Prohaska, S. & Jefferis, G. S. NBLAST: rapid, sensitive comparison of neuronal structure and construction of neuron family databases. Neuron 91, 293–311 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Daniels, R. W., Gelfand, M. V., Collins, C. A. & DiAntonio, A. Visualizing glutamatergic cell bodies and synapses in drosophila larval and adult CNS. J. Comp. Neurol. 508, 131–152 (2008).
Nern, A., Pfeiffer, B. D. & Rubin, G. M. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl Acad. Sci. USA 112, E2967–E2976 (2015).
Li, H. H. et al. A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep. 8, 897–908 (2014).
Selcho, M., Pauls, D., Han, K. A., Stocker, R. F. & Thum, A. S. The role of dopamine in Drosophila larval classical olfactory conditioning. PLoS ONE 4, e5897 (2009).
Selcho, M., Pauls, D., Huser, A., Stocker, R. F. & Thum, A. S. Characterization of the octopaminergic and tyraminergic neurons in the central brain of Drosophila larvae. J. Comp. Neurol. 522, 3485–3500 (2014).
Pauls, D., Selcho, M., Gendre, N., Stocker, R. F. & Thum, A. S. Drosophila larvae establish appetitive olfactory memories via mushroom body neurons of embryonic origin. J. Neurosci. 30, 10655–10666 (2010).
Marley, R. & Baines, R.A. Dissection of third-instar Drosophila larvae for electrophysiological recording from neurons. Cold Spring Harb. Protoc. 2011, pdb.prot065656 (2011).
Acknowledgements
Michael Winding, Mei Shao and Casey Schneider-Mizell contributed equally to this work. We thank Fly Light at HHMI Janelia Research Campus (JRC) for generating confocal images of the GAL4 lines, J. Simpson for sharing a two-photon microscope, Y. Aso and G. Rubin for sharing unpublished driver lines, V. Jayaraman for sharing unpublished versions of CsChrimson and GCamp6f, L. Feng and I. Andrade for help with circuit mapping from EM, K. Hibbard and JRC FLY Core for generating some of the fly stocks, Fly EM at JRC for generating the EM volume, T. Saumweber for discussions, D. Bonnery for help with analyses and Z. Zavala-Ruiz and the JRC Visiting Scientist Program and HHMI JRC for funding. M.Z. was also supported by the European Research Council Consolidator Grant 819650 - LeaRNN, by the Wellcome Trust International Recruitment Supplement 205050/A/16/Z and by the Wellcome Trust Investigator Award 205050/Z/16/Z. C.E was also supported by the European Research Council Consolidator Grant 819650 - LeaRNN and the Wellcome Trust International Recruitment Supplement 205050/A/16/Z. A.C. was supported by Wellcome Trust Grant 205038/Z/16/Z and Wellcome Trust International Recruitment Supplement 205038/A/16/Z. M.W. was also supported by the Wellcome Trust International Recruitment Supplement 205050/A/16/Z and the Wellcome Trust Investigator Award 205050/Z/16/Z. A.L.-K. was supported by the Burroughs Wellcome Foundation, the Gatsby Charitable Foundation, the Simons Collaboration on the Global Brain and NSF award DBI-1707398. B.G. received grant support from the Deutsche Forschungsgemeinschaft (Grant Nos. Ge1091/4-1 and FOR 2705-TP2). A.S.T received grant support from the Deutsche Forschungsgemeinschaft (Grant Nos. TH1584/6-1 and TH1584/7-1).
Author information
Authors and Affiliations
Contributions
C.E. conceived the study, designed and performed experiments and data analysis, and wrote the manuscript. A.F. performed circuit reconstruction and analysis, designed and performed experiments and data analysis, and wrote the manuscript. M.Z. and A.C. conceived the study, performed data analysis, and wrote the manuscript. A.L.-K. developed the model and wrote the manuscript. M.W., C.M.S.-M. and J.V.-A. performed circuit reconstruction and analysis. M.S. performed patch-clamp recordings. R.A., K.E. and T.O. performed experiments. A.S.T. and B.G. contributed data analysis, experimental design and revision of the manuscript. R.D.F. and J.W.T. contributed data and reagents.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Rachel Wilson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Extended Data Fig. 1 Expression patterns of Split-GAL4 lines.
Each panel shows a representative confocal maximum intensity projection (out of N=3) of the complete CNS of third-instar larvae (indicated by the dotted line in the first panel), with the neuropil labeled with anti-N-Cad antibody (blue) and the Split-GAL4line expression pattern revealed with UAS-myr-GFP (green). Arrowheads indicate cell bodies of identified neurons.
Extended Data Fig. 2 Detailed characterization of associative memories formed through different kinds of ‘optogenetic punishments’ or ‘optogenetic rewards’.
Preference scores are shown for the trained odor, ethyl acetate, when it was paired (paired group, closed circles) or not paired (unpaired group, open circles) with optogenetic punishments or rewards. These Preference scores are used to compute the Learning Performance Scores as shown in Fig. 1c,e and 6a-c, additional testing conditions are also shown here. With some natural punishments, aversive memory is behaviorally expressed by trained Drosophila larvae only if the punishment is present at the moment of the test16. Here we assayed olfactory aversive memories in two ways: both with or without optogenetic punishment (red and black bars, respectively) during the retention test. Odor preference was decreased and increased, respectively, relative to genetic controls, after pairing the odor with the presence and absence of the following optogenetic punishments: co-activation of the aversive DAN-f1 and DAN-g1, co-activation of DAN-f1 and DAN-c1, or activation of Basins. Aversive memory formed by DAN activation (green) or by Basins activation (blue) was expressed to the same extent with or without the DANs activated during the retention test. However, memory evoked by the activation of nociceptive MD IV neurons (orange) or FB2N-19 (yellow) was fully expressed only if these neurons were active again during the retention test. Odor preference was increased and decreased, respectively, relative to genetic controls, after pairing the odor with the presence and absence of the following optogenetic rewards: the co-activation of DAN-h1, -i1, and -k1 (dark green); the activation of FB2N-18 and FB2N-11 (yellow), or activation of FAN-7 (blue-gray). Thus, both absence of odor in the unpaired group of animals, as well as the presence of odor in the paired group of animals can be associated with the activation of some DANs or some of their afferent neurons. These results suggest that presenting an odor unpaired with the activation of some of these DANs induces memory of opposite valence to the paired presentation. For other DANs or afferent neurons, only paired (for example A00c, purple) or only unpaired (for example the modulatory DAN-f1, the nociceptive MD IV sensory neuron, or FB2N-19) contingency significantly affected odor preference with respect to the control group. Either of these two observed types of effects can contribute to the negative or positive learning performance indexes plotted in Fig. 1c,e and 6a-c. Sample sizes: N = 42, 11, 17, 16, 12, 14, 12, 13, 12, 16, 12, 14, 12, 12, 15, 14, 12, 11, 14, 13, 12, 14, 11, 11, 11, 18, 11, 20, 25, 33, 52,14, 21, 14, 14, 18, 18, 31, 52, 27, 11,11, 10, 13, 20 (control groups in bold). Mean and standard deviations are shown. Black *: p-value<Holm-Bonferroni-adjusted threshold for 0.05 from a two-sided Wilcoxon signed rank test comparison between paired and unpaired group. Grey *: p-value<Holm-Bonferroni-adjusted threshold for 0.05 from a two-sided Mann-Whitney U test comparison between the preference scores for a given group (paired or unpaired) and the preference scores (for paired or unpaired protocol, respectively) obtained by the control line shown on the left of each set of data. Exact p-values are available in Source Data Fig. 1.
Extended Data Fig. 3 Matrix of similarity between modulator neurons based on the amount of common input.
We computed the cosine similarity of the connectivity matrix of row input neurons onto column modulatory neurons, which is the dot product of the rows, divided by the multiplication of the norm of each row. An input here is a connection, consisting typically of many synapses, from a specific cell type onto the modulatory neuron. Inputs onto a modulatory neuron are considered if the pair of left and right neurons presynaptic to the pair of left and right modulatory neurons is each above a threshold of 1% (for example the presynaptic neuron makes 3 synapses onto a neuron with 300 postsynaptic sites) and the sum of both is over 3.3% (for example the sum of both connections is above 10 synapses for receiving neurons with 300 postsynaptic sites). Interestingly, functionally similar DANs, whose activation leads to aversive memory for paired odors share a higher fraction of presynaptic partners with each other than with other DANs. By contrast some modulatory neurons that innervate the same compartment but express different neurotransmitters (for example OAN-g1 and DAN-g1) receive inputs from drastically different subsets of pre-modulatory neurons. Such modulatory neurons that innervate the same compartment could therefore be differentially recruited during learning. Interestingly though, OAN-e1 and MBIN-e2 in the UVL, whose activation paired with odor did not in-duce memory in our paradigm and that were not significantly activated by fictive punishments share a higher fraction of their input with the VL/LA DANs than with other modulatory neurons. This raises the possibility that the UVL modulatory neurons may be recruited by similar stimuli to the VL/LA DANs, but only in specific circumstances.
Extended Data Fig. 4 Input onto feedback neurons.
Figure shows the fractions of total dendritic input each pre-modulatory neuron (FBN, FB2N or FFN) receives from KCs, modulatory neurons, MBONs, FBNs, FB2Ns, FFNs, and from other non-MB neurons (others). a FBNs receive on average 12% of their inputs directly from MBONs and most of them also receive inputs from other FBNs, with an average of 26% from MBONs and other FBNs combined (see also Supplementary Figure 4a). b FB2Nsreceive inputs both from FBNs (on average 17%) and from other FB2Ns (on average 28% from FBNs and FB2Ns combined).Many feedback neurons also receive a significant fraction of input from other unknown neurons from other brain areas (other than MB), suggesting that the feedback about the learnt valences of stimuli is integrated with or modulated by other information. c Tables show percent of inputs onto FBNs (top) and FB2Ns (bottom) from MBONs, FBNs, FB2Ns, FFNs, modulatory neurons and Kenyon cells.
Extended Data Fig. 5 (related to Fig. 3a) Modulatory neurons receive convergent one-step feedback from multiple MBONs from functionally distinct compartments.
Connectivity of each of the 40 feedback neuron (FBN) pairs that provide one-step feedback from MBONs to DANs. Each diagram represents the connectivity of a single left-right pair of homologous FBNs. Each box indicates a separate compartment. Purple, compartment(s) of the presynaptic MBON(s). Green, compartment(s) of the postsynaptic modulatory neuron(s). FBNs are ordered according to the modulatory neuron they innervate, starting with peduncle modulatory neurons and ending with the medial lobe ones. Classical neurotransmitter profiles of the MBONs and FBNs are indicated by the arrow (cholinergic, excitatory connection), vertical line (GABAergic, inhibitory connection) or square (glutamatergic, probably also inhibitory connection) for the neurons for which they are known from immunostaining (For images, see Extended Data Fig. 9 for FBNs and ref. 12 for MBONs), or by a circle when they are unknown. 7 FBNs provide exclusively within-compartment feedback. 13 FBNs provide exclusively cross-compartment feedback (named Connectivity of each of the 40 feedback neuron (FBN) pairs that provide one-step feedback from MBONs to DANs. Each diagram represents the connectivity of a single left-right pair of homologous FBNs. Each box indicates a separate compartment. Purple, compartment(s) of the presynaptic MBON(s). Green, compartment(s) of the postsynaptic modulatory neuron(s). FBNs are ordered according to the modulatory neuron they innervate, starting with peduncle modulatory neurons and ending with the medial lobe ones. Classical neurotransmitter profiles of the MBONs and FBNs are indicated by the arrow (cholinergic, excitatory connection), vertical line (GABAergic, inhibitory connection) or square (glutamatergic, probably also inhibitory connection) for the neurons for which they are known from immunostaining (For images, see Extended Data Fig. 9 for FBNs and ref.12 for MBONs), or by a circle when they are unknown. 7 FBNs provide exclusively within-compartment feedback. 13 FBNs provide exclusively cross-compartment feedback (named FANs, for feed-across). 8 FBNs synapse onto multiple modulatory neurons from multiple compartments. The largest class of FBNs (17) receives input from multiple MBONs, with the majority (at least 13) receiving input of potentially opposite sign from MBONs from functionally distinct compartments. More than a quarter of FBNs (at least 12) receive direct GABAergic(inhibitory) or glutamatergic (also potentially inhibitory) input from MBONs from one compartment and direct cholinergic(excitatory) input from MBONs from a functionally distinct compartments enabling them to compare the odor drive to these MBONs. Many DANs (DAN-f1, d1, i1, j1, and k1) receive potentially inhibitory (excitatory FBN downstream of an inhibitory MBON) one-step feedback from MBONs from one compartment and potentially disinhibitory (inhibitory FBN downstream of an inhibitory MBON) or excitatory (excitatory FBN downstream of an excitatory MBON) one-step feedback from MBONs from a functionally distinct compartment. A common pattern for the lobe DANs implicated in memory formation may be a likely inhibitory connection from an MBON from their own compartment and a likely disinhibitory connection from an MBON from a compartment of opposite valence (observed for both DAN-g1 and i1), that could enable these DANs to compare the odor drive to MBONs from compartments of opposite valence.
Extended Data Fig. 6 Clustering FBNs based on input from MBONs.
a Heat map of FBN similarity based on the pattern of FBN synaptic inputs from MBONs. The similarity between a pair of FBNs was computed as the cosine similarity between the vectors of their normalized synaptic inputs from all MBONs. Indices were ordered by agglomerative clustering with average linkage (dendrogram shown at top). We highlight six groups of FBNs defined by similarities in their input patterns (bold lines in dendrogram, numbered). b Heat maps showing patterns of input from MBONs onto FBNs for the input groups highlighted in a. In all cases, connectivity is measured in a Heat map of FBN similarity based on the pattern of FBN synaptic inputs from MBONs. The similarity between a pair of FBNs was computed as the cosine similarity between the vectors of their normalized synaptic inputs from all MBONs. Indices were ordered by agglomerative clustering with average linkage (dendrogram shown at top). We highlight six groups of FBNs defined by similarities in their input patterns (bold lines in dendrogram, numbered). b Heat maps showing patterns of input from MBONs onto FBNs for the input groups highlighted in a. In all cases, connectivity is measured in normalized synaptic input on the postsynaptic neuron. Most input groups receive dominant input from a single specific MBON (Groups 1,2, 5) or small group of MBONs (Groups 3 and 4), while Group 6 is not well-clustered and contains a variety of dissimilar input patterns. c Heat maps showing the patterns of synaptic output from FBNs to modulatory neurons for the input groups highlighted in a. d The observed similarity in the output patterns between FBNs within each group, compared to shuffled data. For each group clustered by input pattern, we computed the observed median of cosine similarity of the output vectors across all pairs of neurons (red line). In Groups 1,2, and 3, the neurons clustered by inputs had more similar output patterns than would be expected by chance. To determine significance, we compared the observed similarity to the distribution of the median cosine similarity for randomly permuted samples from the observed population of input vectors (black histograms, n=10000 randomized trials) with a one-sided permutation test. A Holm-Sidak correction was applied to p-values to correct for multiple comparisons. n.s.: p>0.05.
Extended Data Fig. 7 Clustering FBNs based on output onto modulatory neurons.
a Heat map of FBN similarity based on the pattern of FBN synaptic output across all modulatory neurons. The similarity between a pair of FBNs was computed as the cosine similarity between the vectors of normalized synaptic output onto all modulatory neurons. Indices were ordered by agglomerative clustering with average linkage (dendrogram shown at top). We highlight six groups of FBNs defined by similarities in their output patterns (bold lines in dendrogram, numbered). b Heat maps showing patterns of synaptic output from FBNs to modulatory neurons for output groups highlighted in a. Each group corresponds to several FBNs strongly targeting one or a small number of modulatory neurons, suggesting that some modulatory neurons are more strongly modulated than others. c Heat maps showing patterns of input onto FBNs from MBONs for the output groups highlighted in a. d The observed similarity in the input patterns between FBNs within each group, compared to shuffled data. For each group (as defined by output patterns), we computed the observed median of cosine similarity of the input vectors across all pairs of neurons (red line). In Groups 1-5, the neurons clustered by outputs had more input output patterns than would be expected by chance. To determine significance, we compared the observed similarity to the distribution of the median cosine similarity for randomly permuted samples from the observed population of input vectors (black histograms, n=10000 randomized trials) with a one-sided permutation test. A Holm-Sidak correction was applied to p-values to correct for multiple comparisons. n.s.: p>0.05.
Extended Data Fig. 8 Direct MBONs to modulatory neuron connectivity is very sparse, in contrast to the very dense connectivity via one-and two-step feedback pathways.
a Connectivity matrix showing normalized synaptic input (expressed as % input) each modulatory neuron (columns) receives from each MBON (rows). Only reliable connections for which the postsynaptic neuron receives at least 1% of input from the presynaptic neuron are shown. When the neurotransmitter of the MBON is known, the circle is color-coded to represent type of connection: excitatory (ChAT) or probably disinhibitory (GluT). Color shades represent the valence of the memory formed in a given compartment (red: aversive memory, blue: appetitive memory). True within-compartment feedback connections from an MBON that receives direct synaptic input from that modulatory neuron are boxed in bold. Very few modulatory neurons receive direct input from MBONs, in contrast to the dense connectivity between MBONs and modulatory neurons via the indirect one- and two-step feedback pathways (b). b Connectivity matrix showing indirect connections between MBONs and modulatory neurons via one-step and/or two-step feedback pathways. The matrix was obtained by summing the matrices from Fig. 3b and Fig. 5e. The color indicates the type of indirect connection existing between a given MBON and a given DAN. Bubble size represents a connectivity index computed as in Fig. 3b and Fig. 5e. A connectivity index of 1 or 10 means that for all connections comprising that indirect feedback pathway the presynaptic neuron accounts for 1% and 10% of input onto that postsynaptic neuron, respectively. One- and two-step feedback drastically increases the connectivity between MBONs and modulatory neurons, compared to direct connections (a).
Extended Data Fig. 9 Identification of neurotransmitters expressed in some FBNs/FB2Ns.
Neurotransmitter expression detected in neuron somata using antibody labelling. We identified GAL4 lines that drive gene expression in some of the FBN or FB2N neurons and used them to express GFP in these neurons. We stained central nervous systems with antibodies against GFP and either ChAT (choline acetyltransferase), GABA (gamma aminobutyric acid) or GLUT (vesicular glutamate transporter).Each row shows from left to right: the name of the individual neuron, anti-GFP (green), anti-ChAT (magenta), and both antibody stainings combined; anti-GFP (green), anti-GABA (magenta), and both antibody stainings combined; anti-GFP (green) and anti-GLUT (magenta), and both antibody stainings combined. Whether a cell is cholinergic, GABAergic or glutamatergic is listed at the beginning of each row under the neuron name. Images show confocal maximum intensity projections of specific neuronal cell bodies. At least two replicates were obtained per genotype. Scale bars: 5μm.
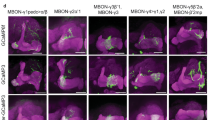
Extended Data Fig. 10 (related to Fig. 6) Identification of driver lines for EM-reconstructed Feedback neurons.
We were able to generate Split-GAL4 lines that drive expression in a single pair of neurons, or in very few cell types, for three different pairs of FBNs or FB2Ns that target VL DANs (a-c). We used these lines to optogenetically activate these neurons instead of a US during an associative learning paradigm (Fig. 6). i) Skeletons of specific feedback neurons reconstructed in the EM. Red dots, presynaptic sites. Blue dots, postsynaptic sites. Grey, mushroom body vertical lobe (MB vl) for reference. d, dendritic arbor. ii) Maximum intensity projections of confocal stacks of larval brains showing the same neurons visualized with reporters targeted using specific Split-GAL4 lines. For some lines multicolor flp-outs were used to visualize each neuron in a different color to facilitate identification and comparison with EM (N=1). Grey, neuropil visualized with N-cad. Dashed line, brain outline. iii) Maximum intensity projections of confocal stacks of the entire nervous system showing the complete expression pattern of each line revealed by driving UAS-myr-GFP. Grey, neuropil visualized with N-cad. Dashed line, nervous system outline. Representative image from N=3. a, The SS02401-Split-GAL4 line drives expression in FB2N-19 (i) in the brain (ii), and very weakly and stochastically (not reproducibly in all samples) in a few ascending neurons and ensheathing glia in the nerve chord (iii). b, The SS02108-Split-GAL4 line drives expression in FAN-7 and MB2ON-86 (i) in the brain visualized with multicolor flp-outs in (ii). Complete expression pattern of SS02108-Split-GAL4 visualized with UAS-myr-GFP shows additional expression in a few somatosensory interneurons in the nerve cord, called ladders, that mediate avoidance behavior and are hence unlikely to have a positive valence and evoke the appetitive memory observed in Fig. 6b. We identified the SS04330-Split-GAL4 line as driving expression specifically in the MB2ON-86 neuron and used it as an additional control in Fig. 6b. c, The SS01778-Split-GAL4 line drives expression in both FB2N-18 and FB2N-11, which have very similar morphology and very similar connectivity (Supplementary Figures 3 and 4b-d). The SS02181-Split-GAL4 line (ii shows multi-color flp-outs) drives expression in FB2N-18 and in MB2IN-207, one of the weakly connected pre-modulatory neurons from lineage DAMv12. Notice the ventrally projecting dendrite (d), a distinctive feature of MB2IN-207 neuron (i). UAS-myr-GFP expression patterns of the two lines show that they do not drive expression in any other neurons in the nerve cord (iii).
Supplementary information
Supplementary Information
Supplementary Table 1, legends for Supplementary Adjacency Matrices 1–3, Atlas, Figs. 1–6.
Supplementary Adjacency Matrix 1
All Connection types. Adjacency matrix between all neurons that are part of the extended mushroom body network (that is, modulatory neurons, MBONs, FFNs, FBNs and FB2Ns), connected via all connection types.
Supplementary Adjacency Matrix 2
Axo-dendritic connections. Adjacency matrix between all neurons that are part of the extended mushroom body network, connected via axo-dendritic synapses.
Supplementary Adjacency Matrix 3
Axo-axonic connections. Adjacency matrix between all neurons that are part of the extended mushroom body network, connected via axo-axonic synapses.
Source data
Rights and permissions
About this article
Cite this article
Eschbach, C., Fushiki, A., Winding, M. et al. Recurrent architecture for adaptive regulation of learning in the insect brain. Nat Neurosci 23, 544–555 (2020). https://doi.org/10.1038/s41593-020-0607-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-0607-9
This article is cited by
-
Optimized design and in vivo application of optogenetically functionalized Drosophila dopamine receptors
Nature Communications (2023)
-
Olfactory navigation in arthropods
Journal of Comparative Physiology A (2023)
-
A single-cell transcriptomic atlas of complete insect nervous systems across multiple life stages
Neural Development (2022)
-
Learning with reinforcement prediction errors in a model of the Drosophila mushroom body
Nature Communications (2021)
-
Dopaminergic mechanism underlying reward-encoding of punishment omission during reversal learning in Drosophila
Nature Communications (2021)