Abstract
Alzheimer’s disease (AD) is characterized by the selective vulnerability of specific neuronal populations, the molecular signatures of which are largely unknown. To identify and characterize selectively vulnerable neuronal populations, we used single-nucleus RNA sequencing to profile the caudal entorhinal cortex and the superior frontal gyrus—brain regions where neurofibrillary inclusions and neuronal loss occur early and late in AD, respectively—from postmortem brains spanning the progression of AD-type tau neurofibrillary pathology. We identified RORB as a marker of selectively vulnerable excitatory neurons in the entorhinal cortex and subsequently validated their depletion and selective susceptibility to neurofibrillary inclusions during disease progression using quantitative neuropathological methods. We also discovered an astrocyte subpopulation, likely representing reactive astrocytes, characterized by decreased expression of genes involved in homeostatic functions. Our characterization of selectively vulnerable neurons in AD paves the way for future mechanistic studies of selective vulnerability and potential therapeutic strategies for enhancing neuronal resilience.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw snRNA-seq sequencing data and unfiltered UMI count matrices are available on the Gene Expression Omnibus (GEO) under accession code GSE147528. Single-cell data after quality control is available for download at Synapse.org under the Synapse ID syn21788402. Post-quality-control data can also be explored interactively through the CellXGene platform at https://kampmannlab.ucsf.edu/ad-brain. Data from Mathys et al.14 was downloaded from Synapse under Synapse ID syn18485175.
Code availability
We provide the full bioinformatics pipeline for the analysis of snRNA-seq data in this paper at https://kampmannlab.ucsf.edu/ad-brain-analysis.
References
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Scholl, M. et al. PET imaging of tau deposition in the aging human brain. Neuron 89, 971–982 (2016).
Seeley, W. W., Crawford, R. K., Zhou, J., Miller, B. L. & Greicius, M. D. Neurodegenerative diseases target large-scale human brain networks. Neuron 62, 42–52 (2009).
Braak, H. & Braak, E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 16, 271–278 (1995).
Price, J. L. et al. Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer disease. Arch. Neurol. 58, 1395–1402 (2001).
Stranahan, A. M. & Mattson, M. P. Selective vulnerability of neurons in layer II of the entorhinal cortex during aging and Alzheimer’s disease. Neural Plast. 2010, 108190 (2010).
Van Hoesen, G. W., Hyman, B. T. & Damasio, A. R. Entorhinal cortex pathology in Alzheimer’s disease. Hippocampus 1, 1–8 (1991).
Gomez-Isla, T. et al. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J. Neurosci. 16, 4491–4500 (1996).
Braak, H. & Braak, E. The human entorhinal cortex: normal morphology and lamina-specific pathology in various diseases. Neurosci. Res. 15, 6–31 (1992).
Kordower, J. H. et al. Loss and atrophy of layer II entorhinal cortex neurons in elderly people with mild cognitive impairment. Ann. Neurol. 49, 202–213 (2001).
Drummond, E. & Wisniewski, T. Alzheimer’s disease: experimental models and reality. Acta Neuropathol. 133, 155–175 (2017).
Lake, B. B. et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016).
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Grubman, A. et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019).
Nelson, P. T. et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J. Neuropathol. Exp. Neurol. 71, 362–381 (2012).
Hof, P. R. et al. Parvalbumin-immunoreactive neurons in the neocortex are resistant to degeneration in Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 50, 451–462 (1991).
Fu, H. et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 22, 47–56 (2019).
Heinsen, H. et al. Quantitative investigations on the human entorhinal area: left-right asymmetry and age-related changes. Anat. Embryol. (Berl.) 190, 181–194 (1994).
Kobro-Flatmoen, A. & Witter, M. P. Neuronal chemo-architecture of the entorhinal cortex: a comparative review. Eur. J. Neurosci. 50, 3627–3662 (2019).
Naumann, R. K. et al. Conserved size and periodicity of pyramidal patches in layer 2 of medial/caudal entorhinal cortex. J. Comp. Neurol. 524, 783–806 (2016).
Ramsden, H. L., Surmeli, G., McDonagh, S. G. & Nolan, M. F. Laminar and dorsoventral molecular organization of the medial entorhinal cortex revealed by large-scale anatomical analysis of gene expression. PLoS Comput. Biol. 11, e1004032 (2015).
Jabaudon, D., Shnider, S. J., Tischfield, D. J., Galazo, M. J. & Macklis, J. D. RORbeta induces barrel-like neuronal clusters in the developing neocortex. Cereb. Cortex 22, 996–1006 (2012).
Oishi, K., Aramaki, M. & Nakajima, K. Mutually repressive interaction between Brn1/2 and Rorb contributes to the establishment of neocortical layer 2/3 and layer 4. Proc. Natl Acad. Sci. USA 113, 3371–3376 (2016).
Nakagawa, Y. & O’Leary, D. D. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev. Neurosci. 25, 234–244 (2003).
Marinaro, F., et al. Molecular and cellular pathology of monogenic Alzheimer’s disease at single cell resolution. Preprint at bioRxiv https://doi.org/10.1101/2020.07.14.202317 (2020).
Liang, W. S. et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol. Genomics 33, 240–256 (2008).
Franjic, D., et al. Molecular diversity among adult human hippocampal and entorhinal cells. Preprint at bioRxiv https://doi.org/10.1101/2019.12.31.889139 (2019).
Ehrenberg, A. J., et al. A manual multiplex immunofluorescence method for investigating neurodegenerative diseases. J. Neurosci. Meth. https://doi.org/10.1016/j.jneumeth.2020.108708. (2020).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of alzheimer’s disease. Cell 169, 1276–1290 e1217 (2017).
Srinivasan, K., et al. Alzheimer’s patient brain myeloid cells exhibit enhanced aging and unique transcriptional activation. Preprint at bioRxiv https://doi.org/10.1101/610345 (2019).
Thrupp, N., et al. Single nucleus sequencing fails to detect microglial activation in human tissue. Preprint at bioRxiv https://doi.org/10.1101/2020.04.13.035386 (2020).
Chen, W. T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991 (2020).
Perez-Nievas, B. G. & Serrano-Pozo, A. Deciphering the Astrocyte reaction in Alzheimer’s disease. Front Aging Neurosci. 10, 114 (2018).
Simpson, J. E. et al. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer’s pathology and APOE genotype. Neurobiol. Aging 32, 1795–1807 (2011).
Sekar, S. et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol. Aging 36, 583–591 (2015).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Laywell, E. D. et al. Enhanced expression of the developmentally regulated extracellular matrix molecule tenascin following adult brain injury. Proc. Natl Acad. Sci. USA 89, 2634–2638 (1992).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Anderson, M. A. et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200 (2016).
Kampmann, M. A CRISPR approach to neurodegenerative diseases. Trends Mol. Med. 23, 483–485 (2017).
Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 16, 465–480 (2020).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-Derived neurons. Neuron 104, 239–255.e212 (2019).
Hof, P. R. & Morrison, J. H. Neocortical neuronal subpopulations labeled by a monoclonal antibody to calbindin exhibit differential vulnerability in Alzheimer’s disease. Exp. Neurol. 111, 293–301 (1991).
Hof, P. R., Cox, K. & Morrison, J. H. Quantitative analysis of a vulnerable subset of pyramidal neurons in Alzheimer’s disease: I. Superior frontal and inferior temporal cortex. J. Comp. Neurol. 301, 44–54 (1990).
Mikkonen, M., Alafuzoff, I., Tapiola, T., Soininen, H. & Miettinen, R. Subfield- and layer-specific changes in parvalbumin, calretinin and calbindin-D28K immunoreactivity in the entorhinal cortex in Alzheimer’s disease. Neuroscience 92, 515–532 (1999).
Montine, T. J. et al. National Institute on Aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 123, 1–11 (2012).
Hughes, C. P., Berg, L., Danziger, W. L., Coben, L. A. & Martin, R. L. A new clinical scale for the staging of dementia. Br. J. Psychiatry 140, 566–572 (1982).
Grinberg, L. T. et al. Brain bank of the brazilian aging brain study group - a milestone reached and more than 1,600 collected brains. Cell Tissue Bank 8, 151–162 (2007).
Hyman, B. T. et al. National institute on Aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 8, 1–13 (2012).
Suemoto, C. K. et al. Neuropathological diagnoses and clinical correlates in older adults in Brazil: A cross-sectional study. PLoS Med. 14, e1002267 (2017).
Cairns, N. J. et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the consortium for frontotemporal lobar degeneration. Acta Neuropathol. 114, 5–22 (2007).
Ferrer, I., Santpere, G. & van Leeuwen, F. W. Argyrophilic grain disease. Brain 131, 1416–1432 (2008).
Rodriguez, R. D. & Grinberg, L. T. Argyrophilic grain disease: An underestimated tauopathy. Dement. Neuropsychol. 9, 2–8 (2015).
Rodriguez, R. D. et al. Argyrophilic grain disease: Demographics, clinical, and neuropathological features from a large autopsy study. J. Neuropathol. Exp. Neurol. 75, 628–635 (2016).
Braak, H., Thal, D. R., Ghebremedhin, E. & Del Tredici, K. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J. Neuropathol. Exp. Neurol. 70, 960–969 (2011).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Tabula Muris, C. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Lun, A. T., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Stuart, T. et al. Comprehensive Integration of Single-Cell data. Cell 177, 1888–1902 e1821 (2019).
Johansen, N. & Quon, G. scAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol. 20, 166 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Amezquita, R. A. et al. Orchestrating single-cell analysis with bioconductor. Nat. Methods 17, 137–145 (2020).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003).
Stelzer, G. et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 54, 13031–313033 (2016).
Arriza, J. L. et al. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J. Neurosci. 14, 5559–5569 (1994).
Borden, L. A. et al. Cloning of the human homologue of the GABA transporter GAT-3 and identification of a novel inhibitor with selectivity for this site. Receptors Channels 2, 207–213 (1994).
Gendreau, S. et al. A trimeric quaternary structure is conserved in bacterial and human glutamate transporters. J. Biol. Chem. 279, 39505–39512 (2004).
Häberle, J. et al. Congenital glutamine deficiency with glutamine synthetase mutations. N. Engl. J. Med. 353, 1926–1933 (2005).
Kawakami, H., Tanaka, K., Nakayama, T., Inoue, K. & Nakamura, S. Cloning and expression of a human glutamate transporter. Biochem. Biophys. Res. Commun. 199, 171–176 (1994).
Melzer, N., Biela, A. & Fahlke, C. Glutamate modifies ion conduction and voltage-dependent gating of excitatory amino acid transporter-associated anion channels. J. Biol. Chem. 278, 50112–50119 (2003).
Südhof, T. C. Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell 171, 745–769 (2017).
Pellissier, F., Gerber, A., Bauer, C., Ballivet, M. & Ossipow, V. The adhesion molecule Necl-3/SynCAM-2 localizes to myelinated axons, binds to oligodendrocytes and promotes cell adhesion. BMC Neurosci. 8, 90 (2007).
González-Castillo, C., Ortuño-Sahagún, D., Guzmán-Brambila, C., Pallàs, M. & Rojas-Mayorquín, A. E. Pleiotrophin as a central nervous system neuromodulator, evidences from the hippocampus. Front. Cell Neurosci. 8, 443 (2014).
Siddiqui, T. J. et al. An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron 79, 680–695 (2013).
Heinsen, H., Arzberger, T. & Schmitz, C. Celloidin mounting (embedding without infiltration) - a new, simple and reliable method for producing serial sections of high thickness through complete human brains and its application to stereological and immunohistochemical investigations. J. Chem. Neuroanat. 20, 49–59 (2000).
Insausti, R. & Amaral, D. G. Entorhinal cortex of the monkey: IV. Topographical and laminar organization of cortical afferents. J. Comp. Neurol. 509, 608–641 (2008).
Insausti, R., Munoz-Lopez, M., Insausti, A. M. & Artacho-Perula, E. The human periallocortex: layer pattern in presubiculum, parasubiculum and entorhinal cortex. A Review. Front. Neuroanat. 11, 84 (2017).
Rose, S. Vergleichende messungen im allocortex bei tier und mensch. J. Psychol. Neurol. 34, 250–255 (1927).
Fu, H. et al. Tau pathology induces excitatory neuron loss, grid cell dysfunction, and spatial memory deficits reminiscent of early Alzheimer’s disease. Neuron 93, 533–541.e535 (2017).
Hevner, R. F. et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001).
Ferrari, S. L. P. & Cribari-Neto, F. Beta regression for modelling rates and proportions. J. Appl. Stat. 31, 799–815 (2004).
Acknowledgements
We thank A. Pisco, A. Maynard, S. Darmanis and the MACA team at the Chan Zuckerberg Biohub for advice on analysis 10X library preparation and reagents. We thank members of the Kampmann laboratory (A. Samelson, X. Guo, R. Tian and B. Rooney) for feedback on the manuscript. This work was supported by National Institutes of Health (NIH) awards F30 AG066418 (to K.L.), K08 AG052648 (to S.S.), R56 AG057528 (to M.K. and L.T.G.), K24 AG053435 (to L.T.G.), U54 NS100717 (to L.T.G. and M
Author information
Authors and Affiliations
Contributions
K.L., E.L., L.T.G. and M.K. conceptualized and led the overall project. K.L., L.T.G. and M.K. wrote the manuscript, with input from all co-authors. K.L. analyzed snRNA-Seq data and visualized results. E.L. generated snRNA-Seq data, with support from R.S., M.T., and N.N. R.D.R., C.K.S., R.E.P.L., A.E., C.A.P. W.W.S., and S.S. contributed to neuropathological data generation and analysis, R.E., A.P. and H.H. contributed to neuropathological data analysis, and S.H.L. contributed to neuropathological method development.
Corresponding authors
Ethics declarations
Ethics Declarations
This project was approved the ethical committee of the University of Sao Paulo (for tissue transfer) and deemed non-human subject research by UCSF.
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Asgeir Kobro-Flatmoen, Menno Witter, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
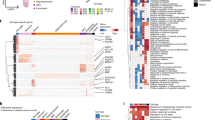
Extended Data Fig. 1 Data quality and initial clustering without cross-sample alignment.
a-b, Mean number of genes (a) or UMIs (b) detected per cell across individual samples for major cell types identified in each dataset. Grubman et al.15 did not resolve excitatory neurons from inhibitory neurons. Pericytes were identified only in Mathys et al.14 Cell type abbreviations: Exc – excitatory neurons, Oligo – oligodendrocytes, Astro – astrocytes, Inh – inhibitory neurons, OPC – oligodendrocyte precursor cells, Micro – microglia, Endo – endothelial cells, Per – pericytes. c-d, tSNE projection of cells from the EC (c) and SFG (d) clustered without first performing cross-sample alignment, colored by individual of origin (center) or cluster assignment (outer). e-f, Heatmap and hierarchical clustering of clusters and cluster marker expression (top subpanels); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Expression of cell type markers (bottom subpanels).
Extended Data Fig. 2 Expression of selected EC excitatory neuron subpopulation markers and pathway enrichment analysis of differentially expressed genes in selectively vulnerable EC excitatory neuron subpopulations.
a, Expression heatmap of genes that are specifically expressed by four or fewer EC excitatory neuron subpopulations; “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). b-d, Enrichment analysis against Gene Ontology Cellular Component terms or Reactome Pathways (b,d) and functional association network analysis (c,e; see Methods) of genes with higher (b-c) or lower expression (d-e) in RORB+ vulnerable EC excitatory neurons, with selected terms highlighted by color. In panels c and e, genes with stronger associations are connected by thicker lines, and genes without known associations are not shown.
Extended Data Fig. 3 Differential expression analysis across Braak stages for EC excitatory neuron subpopulations.
a-b, Number of differentially expressed genes in EC excitatory neuron subpopulations with higher (a) or lower (b) expression in Braak stage 6 vs. Braak stage 0. c-f, Enrichment analysis against Gene Ontology Cellular Component terms (c-d) or Reactome Pathways (e-f) of differentially expressed genes in EC excitatory neuron subpopulations with higher (c,e) or lower (d,f) expression in Braak stage 6 vs. Braak stage 0.
Extended Data Fig. 4 Alignment of EC and SFG maps homologous excitatory neuron subpopulations.
a, tSNE projection of excitatory neurons from the EC and SFG in the joint alignment space, colored by subpopulation identity (top), individual of origin (middle), or brain region (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations across Braak stages (second and third subpanels); for each brain region, statistical significance of differences in relative abundance across Braak stages (Braak 0 n = 3, Braak 2 n = 4, Braak 6 n = 3, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of EC layer-specific genes identified from Ramsden et al.22 (fourth subpanel). Expression heatmap of neocortical layer-specific genes from Lake et al.12 (fifth subpanel). Expression of selectively vulnerable EC excitatory neuron subpopulation markers by excitatory neurons in the EC (sixth subpanel) or SFG (bottom subpanel). Significant beta regression P values (adjusted for multiple testing) are shown in a table at the bottom of the panel. c, Sankey diagram connecting subpopulation identity of excitatory neurons in the EC alignment space and the SFG alignment space to subpopulation identity in the EC+SFG alignment space. The links connecting EC:Exc.s2 and EC:Exc.s4 to SFG:Exc.s2 and SFG:Exc.s4, respectively, are highlighted.
Extended Data Fig. 5 Cross-sample alignment of excitatory neurons from Mathys et al. recapitulates selective vulnerability in a RORB-expressing subpopulation.
a, tSNE projection of excitatory neurons from Mathys et al.14 in the alignment space, colored by subpopulation identity (top) or individual of origin (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations in in AD cases vs. controls, separated by sex (second and third subpanels); for each sex, statistical significance of differences in relative abundance between AD cases vs. controls (cases n = 12, controls n=12, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of neocortical layer-specific genes from Lake et al.12 (fourth subpanel). Expression of selectively vulnerable EC excitatory neuron subpopulation markers (bottom subpanel). c, Heatmap of Pearson correlation between the gene expression profiles of excitatory neuron subpopulations from the EC vs. those from the prefrontal cortex in Mathys et al.14.
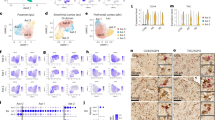
Extended Data Fig. 6 Delineation of the EC for each case used in immunofluorescence validation.
a, The borders of the caudal EC delineated on sections stained with hematoxylin and eosin (H&E) for all 26 cases used in immunofluorescence validation (Table 1). b, Borders of the EC were determined with the aid of 400 um thick serial coronal sections of whole-brain hemispheres stained with gallocyanin (see Methods). Each H&E section (left) along with its corresponding immunofluorescence image (middle) was aligned to the most approximate gallocyanin section (right), in which the the dissecans layers (diss-1, diss-2, and diss-ext) characteristic of the caudal EC were easier to visualize. This was then used to guide delineation of the EC on the H&E and immunofluorescence sections. For more details on the cytoarchitectonic definitions used to define the caudal EC, please consult Heinsen et al.19.
Extended Data Fig. 7 Inhibitory neurons from Mathys et al. also do not show differences in resilience or vulnerability to AD.
a, tSNE projection of inhibitory neurons from Mathys et al.14 in the alignment space, colored by subpopulation identity (top) or individual of origin (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation markers (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations in in AD cases vs. controls, separated by sex (second and third subpanels); for each sex, statistical significance of differences in relative abundance between AD cases vs. controls (cases n=12, controls n=12, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of inhibitory neuron subtype markers from Lake et al.12 (bottom subpanel).
Extended Data Fig. 8 Subclustering of microglia does not sufficiently resolve disease associated microglia signature.
a-c, tSNE projection of astrocytes from the EC (a), SFG (b), and Mathys et al.14 (c) in their respective alignment spaces, colored by subpopulation identity (left) or individual of origin (right). d-f, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanels); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations (middle subpanels) across Braak stages in the EC and SFG (for each brain region, Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) or between AD cases vs. controls in Mathys et al.14 (for each sex, cases n =12, controls n = 12, where n is the number of individuals sampled); statistical significance of differences in relative abundance was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression of disease associated microglia markers, with median expression level marked by line (bottom subpanels).
Extended Data Fig. 9 Subclustering of oligodendrocytes identifies subpopulations with higher expression of AD-associated oligodendrocyte markers from Mathys et al.
a-c, tSNE projection of oligodendrocytes from the EC (a), SFG (b), and Mathys et al.14 (c) in their respective alignment spaces, colored by subpopulation identity (left) or individual of origin (right). d-f, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanels); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations (middle subpanels) across Braak stages in the EC and SFG (for each brain region, Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) or between AD cases vs. controls in Mathys et al.14 (for each sex, cases n =12, controls n = 12, where n is the number of individuals sampled); statistical significance of differences in relative abundance was determined by beta regression and adjusted for multiple comparisons (see Methods). Relative expression of AD-associated oligodendrocyte subpopulation markers from Mathys et al.14 (bottom subpanels).
Extended Data Fig. 10 Astrocyte subpopulations with high GFAP expression from Mathys et al. are highly similar to those from the EC and SFG.
a, tSNE projection of astrocytes from Mathys et al.14 in the alignment subspace, colored by subpopulation identity (top) or individual of origin (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations in in AD cases vs. controls, separated by sex (middle subpanels); for each sex, statistical significance of differences in relative abundance between AD cases vs. controls (cases n=12, controls n=12, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression of genes associated with reactive astrocytes, with median expression level marked by line (bottom subpanel). c, Enrichment analysis of overlap between differentially expressed genes in astrocytes with high GFAP expression from Mathys et al.14 vs. differentially expressed genes in astrocytes with high GFAP expression from the EC and SFG; the number of genes in each gene set and the number of overlapping genes are shown in parentheses, and the hypergeometric test p-values are shown without parentheses.
Supplementary information
Supplementary Tables
Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Leng, K., Li, E., Eser, R. et al. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat Neurosci 24, 276–287 (2021). https://doi.org/10.1038/s41593-020-00764-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00764-7
This article is cited by
-
The concept of resilience to Alzheimer’s Disease: current definitions and cellular and molecular mechanisms
Molecular Neurodegeneration (2024)
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Data-driven modelling of neurodegenerative disease progression: thinking outside the black box
Nature Reviews Neuroscience (2024)
-
Microglia regulation of central nervous system myelin health and regeneration
Nature Reviews Immunology (2024)
-
Single-nucleus RNA velocity reveals critical synaptic and cell-cycle dysregulations in neuropathologically confirmed Alzheimer’s disease
Scientific Reports (2024)